To the editor:
Acute myeloid leukemia (AML) is an aggressive malignancy with a variety of genetic and epigenetic aberrations pinpointing a multistep process of leukemogenesis.1 It is hierarchically organized, with bulk leukemic cells derived from leukemia-initiating cells that possess self-renewal capacity and are capable of establishing leukemia in vivo.2 Recently, preleukemic stem cells (preLSCs) have been described in AML. These clonal cells, derived from hematopoietic stem and progenitor cells (HSPCs), initiate the leukemogenic process but retain their ability to differentiate into mature blood cells. Importantly, preLSCs can survive chemotherapy, suggesting that they constitute a reservoir for leukemia recurrence.3-5
TP53 is an essential tumor suppressor gene located on chromosome 17p13.1. Germ line TP53 mutations characterize the Li-Fraumeni syndrome and occur at low frequencies in patients with AML.6,7 Somatic TP53 mutations have been detected in up to 20% of AMLs, often associated with a complex karyotype. Most importantly, patients with AML with TP53 mutations show resistance to intensive treatments, with inferior survival rates.8-10 On the basis of these facts, “AML with TP53 mutations, chromosomal aneuploidy, or both” has been proposed as a distinct AML subtype, and testing for TP53 mutations has been incorporated into the 2017 European LeukemiaNet recommendations.1,11
We hypothesized that somatic TP53 mutations are early leukemogenic events. By transforming HSPCs into preLSCs, TP53 mutations contribute substantially to the development of AML and its therapeutic resistance.
The study was approved by the ethical committee of the Medical University of Graz (Graz, Austria). Initially, we analyzed 150 diagnostic AML specimens for TP53 mutations (methods described in the supplemental Data, available on the Blood Web site).6,7 Thirty-nine somatic TP53 mutations were detected in 32 specimens (missense, 29 [74%]; nonsense, 8 [21%]; and splice site, 2 [5%]), and loss of the wild-type allele was observed in 5 (16%) of 32 (supplemental Table 1; supplemental Figure 1).
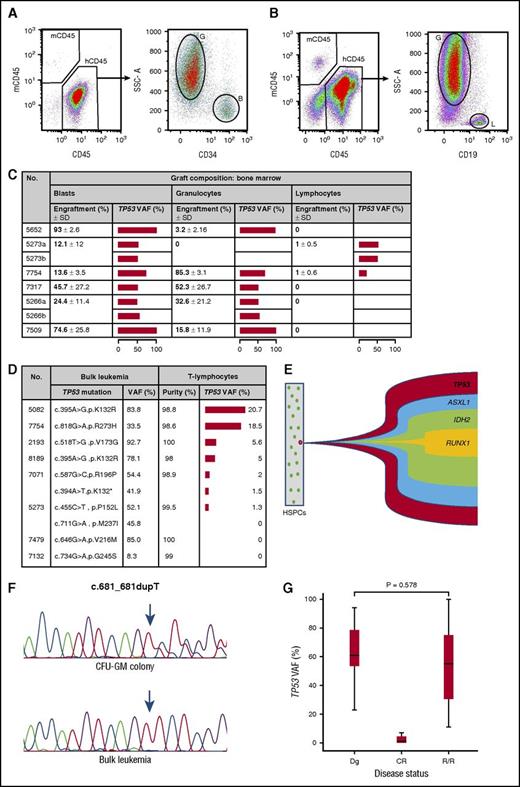
To gain insight into the properties of ancestral cells affected by somatic TP53 mutations, a NOD-scid IL2rγnull mouse model engineered to express human stem-cell factor, granulocyte-macrophage colony-stimulating factor and interleukin-3 (NSGS mice) was employed (data supplement).12 Unpurified bulk leukemia cells of 6 TP53-mutant AMLs were injected into 5 to 6 mice each, which were analyzed 12 to 16 weeks thereafter. The mean human CD45+ engraftment rate per AML specimen was between 31% and 63% in bone marrow and 4% and 52% in spleen, respectively (supplemental Figure 2A-B). A majority of cells engrafted revealed a blast phenotype. Nevertheless, differentiation into granulocytes was observed in 83% of specimens in the bone marrow and 67% in the spleen. Differentiation into B lymphocytes occurred in 33% of specimens in the bone marrow and 17% in the spleen (Figure 1A-B). Importantly, in all human cell types engrafted, the patient-specific somatic TP53 mutation was detected in up to 100% as assessed by ultradeep sequencing (Figure 1C; supplemental Figure 2C; supplemental Table 2). To corroborate these findings, highly purified T lymphocytes obtained from patients at AML diagnosis were analyzed (data supplement). As shown in Figure 1D, a median purity of 99.2% (range, 98% to 100%) of CD45+/CD3+ cells was obtained. Using targeted deep sequencing, the leukemia-specific TP53 mutation was detected in 75% of specimens at a median variant allele frequency of 5% (range, 1.3% to 20.7%). These data indicate that somatic TP53 mutations in AML affect preLSCs that retain their ability to differentiate into mature blood cells in both experimental animals and patients with AML. They support a role for TP53 mutations as early events of acute myeloid leukemogenesis. A number of mutated genes in preLSCs of patients with AML have been described to date, including DNMT3A, TET2, and IDH1/2.4,5 Interestingly, these genes act as epigenetic modifiers, whereas one of the fundamental roles of p53 is related to cell-cycle control, DNA repair, and apoptosis. Dysregulation of these pivotal functions might be an alternative mechanism in establishing a proleukemogenic state in HSPCs.13
In AML, somatically acquired TP53 mutations characterize preleukemic stem cells, are initiating genetic events, and mediate resistant disease. (A) FACS analysis of bone marrow of an NSGS mouse engrafted with 1 × 106 unpurified, human TP53-mutated AML cells showing both blast cells as well as maturation into granulocytes. (B) Engrafted human cells with a blast-cell and B-lymphocyte phenotype. (C) Graft composition of mouse bone marrow. Human blasts were characterized by a sideward scatter (SSC) low/CD34+/CD45dim/CD19− phenotype, granulocytes by a SSChigh/CD33+/CD34−/CD19−, and B lymphocytes by an SSClow/CD34−/CD33−/CD19+ phenotype. The horizontal bar depicts mean TP53 variant allele frequencies (VAFs). (D) Highly purified peripheral blood CD45+/CD3+ cells were obtained at AML diagnosis, and the TP53 VAFs were assessed using ultradeep sequencing. Samples 7071 and 5273, respectively, exhibited 2 different somatic TP53 mutations. Note that in each case analyzed and scored positive, the TP53 VAF exceeded the minute impure fraction of sorted T lymphocytes, thereby excluding results biased because of contamination of AML cells. (E) Data obtained from colony-forming unit–granulocyte, monocyte (CFU-GM) colonies derived from specimen 5652 revealed the TP53 mutation as the initiating event (positive in 38 of 38 colonies), followed by an ASXL1 mutation (37 of 38), IDH2 mutation (20 of 38), and RUNX1 mutation (19 of 38). All cooperating mutations developed sequentially in the TP53-mutated clone. The exact mutation type is shown in Table 1. (F) Loss of heterozygosity at the TP53 locus of samples from UPN 7317. In CFU-GM colonies, a heterozygous TP53 c.681_681dupT mutation is shown, whereas in bulk leukemia cells, the wild-type allele was lost, resulting in a hemizygous state. (G) Quantitative assessment of the TP53 mutational load by the ultradeep sequencing, indicating comparable levels between diagnostic specimens and those obtained at relapsed or refractory (R/R) phase (P = .578 by the exact permutation test for related samples). B, blast cells; CR, complete remission; Dg, diagnosis; G, granulocytes; h, human; L, lymphocytes; m, mouse; SD, standard deviation.
In AML, somatically acquired TP53 mutations characterize preleukemic stem cells, are initiating genetic events, and mediate resistant disease. (A) FACS analysis of bone marrow of an NSGS mouse engrafted with 1 × 106 unpurified, human TP53-mutated AML cells showing both blast cells as well as maturation into granulocytes. (B) Engrafted human cells with a blast-cell and B-lymphocyte phenotype. (C) Graft composition of mouse bone marrow. Human blasts were characterized by a sideward scatter (SSC) low/CD34+/CD45dim/CD19− phenotype, granulocytes by a SSChigh/CD33+/CD34−/CD19−, and B lymphocytes by an SSClow/CD34−/CD33−/CD19+ phenotype. The horizontal bar depicts mean TP53 variant allele frequencies (VAFs). (D) Highly purified peripheral blood CD45+/CD3+ cells were obtained at AML diagnosis, and the TP53 VAFs were assessed using ultradeep sequencing. Samples 7071 and 5273, respectively, exhibited 2 different somatic TP53 mutations. Note that in each case analyzed and scored positive, the TP53 VAF exceeded the minute impure fraction of sorted T lymphocytes, thereby excluding results biased because of contamination of AML cells. (E) Data obtained from colony-forming unit–granulocyte, monocyte (CFU-GM) colonies derived from specimen 5652 revealed the TP53 mutation as the initiating event (positive in 38 of 38 colonies), followed by an ASXL1 mutation (37 of 38), IDH2 mutation (20 of 38), and RUNX1 mutation (19 of 38). All cooperating mutations developed sequentially in the TP53-mutated clone. The exact mutation type is shown in Table 1. (F) Loss of heterozygosity at the TP53 locus of samples from UPN 7317. In CFU-GM colonies, a heterozygous TP53 c.681_681dupT mutation is shown, whereas in bulk leukemia cells, the wild-type allele was lost, resulting in a hemizygous state. (G) Quantitative assessment of the TP53 mutational load by the ultradeep sequencing, indicating comparable levels between diagnostic specimens and those obtained at relapsed or refractory (R/R) phase (P = .578 by the exact permutation test for related samples). B, blast cells; CR, complete remission; Dg, diagnosis; G, granulocytes; h, human; L, lymphocytes; m, mouse; SD, standard deviation.
Next, we determined cooperating genetic aberrations of TP53-mutated AMLs. Bulk leukemia cells were analyzed by whole-exome sequencing and targeted deep sequencing, respectively. Cooperating mutations identified were then assessed in CFU-GM colonies derived from sorted Lin-CD34+/CD38−/CD99− single cells (data supplement; supplemental Figure 3A). The patient-specific TP53 mutation was present in the vast majority of colonies (median, 97%; range, 45% to 100%); however, only a paucity of cooperating mutations in cancer gene census genes was detected (median, 1; range, 0-3). Some of them (DNMT3A, IDH2, RUNX1) have been described as early events in AML before. Notably, in all specimens analyzed, cooperating mutations developed sequentially or concomitantly in the TP53-mutated clone (Table 1; supplemental Figure 3C). Whole-exome sequencing revealed an abundance of copy-number alterations, with a median of 37 chromosomal losses (range, 27-80) and 34 chromosomal gains (range, 21-113) per sample (Table 1; supplemental Figure 3B). The fact that loss of heterozygosity at the TP53 locus was shown in bulk leukemia cells but not in CFU-GM colonies (Figure 1F) supports previous concepts of copy-number alterations as secondary events after the onset of TP53 mutations.14,15 From 20 patients with secondary or therapy-related AML exhibiting somatic TP53 mutations, material from antecedent hematological disorders was available. Using Sanger and targeted deep sequencing, respectively, the particular TP53 variant could be shown in 18 of them (90%; supplemental Table 3). These data showing TP53 mutations as initiating events are in line with reports on therapy-related AML indicating that the leukemia-specific TP53 aberration was already present at low levels in normal bone marrow before commencement of cytotoxic treatments for the primary malignancy.16,17 Using a mouse model with bone marrow chimeric wild-type and TP53+/− HSPCs, Wong et al17 further demonstrated that haploinsufficient p53 cells preferentially expanded after cytotoxic exposure. Recently, clinical studies showed that patients with a primary malignancy and clonal hematopoiesis of undeterminate potential (CHIP) at the time of initial antineoplastic treatment and autologous stem cell transplantation, respectively, are at increased risk of therapy-related myeloid neoplasms as compared with those without CHIP. TP53 mutations were frequently detected clonal aberrations in these patients, with the mutant clone expanding substantially over time.18-20 Furthermore, TP53 mutations are among those found in healthy individuals with CHIP who also exhibited an increased risk of developing various blood cancers.21,22
Analysis of concomitant genetic aberrations in AML with somatic TP53 mutations
| No. . | TP53 VAF, % . | TP53 and concomitant mutations . | No. of mutated colonies (%) . | Copy number alterations . | ||||
|---|---|---|---|---|---|---|---|---|
| Losses . | Gains . | |||||||
| No. . | Length, bp . | No. . | Length, bp . | . | ||||
| 7549 | 65.9 | TP53: p.E221Gfs*26 | 44 of 60 (73) | 28 | 285.789.803 | 34 | 32.999.479 | |
| SRSF2: p.P95L | 38 of 60 (63) | |||||||
| BCL11A: p.P702L | 26 of 60 (43) | |||||||
| 5266 | 44.0 | TP53: p.L195P | 10 of 11 (91) | 80 | 402.787.349 | 21 | 9.619.914 | |
| ATP1A1: p.N483S | 6 of 11 (55) | |||||||
| TP53: p.Q165* | 5 of 11 (45) | |||||||
| 5273 | 52.0 | TP53: p.M237I | 107 of 112 (96) | 36 | 572.663.152 | 34 | 58.851.203 | |
| TP53: p.P152L | 106 of 112 (95) | |||||||
| 7317 | 88.1 | TP53: p.Asp228* | 18 of 18 (100) | 52 | 367.103.483 | 45 | 87.828.354 | |
| 7509 | 64.0 | TP53: p.V157F | 67 of 67 (100) | 38 | 182.926.187 | 30 | 55.245.921 | |
| DNMT3A: p.Q886R | 67 of 67 (100) | |||||||
| 7071 | 54.4 | TP53: p.R196P | 62 of 62 (100) | 27 | 131.583.940 | 113 | 316.102.979 | |
| TP53: p.K132* | 60 of 62 (97) | |||||||
| FLT3: p.D835H | 57 of 62 (92) | |||||||
| PTPN11: p.D61Y | 2 of 62 (3) | |||||||
| 5652 | 89.9 | TP53: p.R175H | 38 of 38 (100) | NA | NA | NA | NA | |
| ASXL1: p.G658* | 37 of 38 (97) | |||||||
| IDH2: p.R140Q | 20 of 38 (53) | |||||||
| RUNX1: p.V179C fs* 34 | 19 of 38 (50) | |||||||
| 7139 | 51.9 | TP53: p.I255N | 55 of 55 (100) | NA | NA | NA | NA | |
| No. . | TP53 VAF, % . | TP53 and concomitant mutations . | No. of mutated colonies (%) . | Copy number alterations . | ||||
|---|---|---|---|---|---|---|---|---|
| Losses . | Gains . | |||||||
| No. . | Length, bp . | No. . | Length, bp . | . | ||||
| 7549 | 65.9 | TP53: p.E221Gfs*26 | 44 of 60 (73) | 28 | 285.789.803 | 34 | 32.999.479 | |
| SRSF2: p.P95L | 38 of 60 (63) | |||||||
| BCL11A: p.P702L | 26 of 60 (43) | |||||||
| 5266 | 44.0 | TP53: p.L195P | 10 of 11 (91) | 80 | 402.787.349 | 21 | 9.619.914 | |
| ATP1A1: p.N483S | 6 of 11 (55) | |||||||
| TP53: p.Q165* | 5 of 11 (45) | |||||||
| 5273 | 52.0 | TP53: p.M237I | 107 of 112 (96) | 36 | 572.663.152 | 34 | 58.851.203 | |
| TP53: p.P152L | 106 of 112 (95) | |||||||
| 7317 | 88.1 | TP53: p.Asp228* | 18 of 18 (100) | 52 | 367.103.483 | 45 | 87.828.354 | |
| 7509 | 64.0 | TP53: p.V157F | 67 of 67 (100) | 38 | 182.926.187 | 30 | 55.245.921 | |
| DNMT3A: p.Q886R | 67 of 67 (100) | |||||||
| 7071 | 54.4 | TP53: p.R196P | 62 of 62 (100) | 27 | 131.583.940 | 113 | 316.102.979 | |
| TP53: p.K132* | 60 of 62 (97) | |||||||
| FLT3: p.D835H | 57 of 62 (92) | |||||||
| PTPN11: p.D61Y | 2 of 62 (3) | |||||||
| 5652 | 89.9 | TP53: p.R175H | 38 of 38 (100) | NA | NA | NA | NA | |
| ASXL1: p.G658* | 37 of 38 (97) | |||||||
| IDH2: p.R140Q | 20 of 38 (53) | |||||||
| RUNX1: p.V179C fs* 34 | 19 of 38 (50) | |||||||
| 7139 | 51.9 | TP53: p.I255N | 55 of 55 (100) | NA | NA | NA | NA | |
Concomitant mutations were initially determined by whole-exome sequencing and targeted deep sequencing (samples 5652 and 7139), respectively, of bulk leukemia cells and then assessed in CFU-GM colonies derived from sorted Lin-CD34+/CD38−/CD99− single cells. A total of 423 colonies were assessed, with a median of 58.5 colonies per samples (range, 11-112). Copy-number alterations were assessed by whole-exome sequencing of bulk leukemia cells. The length of gains and losses refers to the total number of alterations observed per sample.
NA, not available; VAF, variant allele frequency of TP53 as determined by targeted deep sequencing of diagnostic specimens.
Finally, we assessed the role of somatic TP53 mutations with respect to resistant disease (data supplement). When analyzing 59 patients who received intensive treatments for their AML (supplemental Table 4), the estimated 5-year overall survival rates for TP53 wild-type and TP53-mutated cases were 18% and 0% (P = .008), and the 5-year event-free survival rates were 16% and 0% (P = .033), respectively (supplemental Figure 4A-B), confirming previous reports.8-10 Ultradeep sequencing of sequential bone marrow specimens from patients with TP53-mutated AML at the time of diagnosis, complete remission, and relapsed or refractory disease revealed a decrease of the median TP53 variant allele frequency from 67.5% at diagnosis to 1% at complete remission. However, at relapsed or refractory stages, it rose again to 45.5%, comparable to diagnostic levels (P = .578; Figure 1G; supplemental Table 5).
In summary, we show that somatic TP53 mutations characterize preLSCs in AML, using both a xenograft mouse model and primary AML specimens. TP53 mutations represent initiating mutations in this type of leukemia and are mediators of resistant disease in AML. These data add further evidence to recent claims of TP53-mutated AML as a distinct disease entity and have implications for the development of targeted treatment approaches.
The online version of the article contains a data supplement.
Authorship
Acknowledgments: This work was supported by the PhD program of the Medical University of Graz, Graz, Austria (R.L.), Leukämiehilfe Steiermark, and unrestricted grants from Teva, Celgene, and Gerot Lannacher.
Contribution: H.S. designed and supervised the study; R.L., K.L., E.H., P.U., K.A., K.K., E.S., A.R., B.F., S.H., B.R., V.S., M.G.S., A.Z., A.W., and H.S. acquired data; J.M.M., C.T., F.G.R., and K.D. provided patient samples and clinical data; R.L., K.L., E.H., P.U., K.K., E.S., V.S., M.G.S., G.H., K.D., A.Z., A.W., and H.S. analyzed and interpreted data; R.L. and H.S. wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: H.S. received grant support from Teva, Celgene, and Gerot Lannacher; C.T. is part owner of AgenDix GmbH. The remaining authors declare no competing financial interests.
Correspondence: Heinz Sill, Division of Hematology, Medical University of Graz, Auenbruggerplatz 38, A-8036 Graz, Austria; e-mail: heinz.sill@medunigraz.at.