Abstract
The genetic landscape of classical myeloproliferative neoplasm (MPN) is in large part elucidated. The MPN-restricted driver mutations, including those in JAK2, calreticulin (CALR), and myeloproliferative leukemia virus (MPL), abnormally activate the cytokine receptor/JAK2 pathway and their downstream effectors, more particularly the STATs. The most frequent mutation, JAK2V617F, activates the 3 main myeloid cytokine receptors (erythropoietin receptor, granulocyte colony-stimulating factor receptor, and MPL) whereas CALR or MPL mutants are restricted to MPL activation. This explains why JAK2V617F is associated with polycythemia vera, essential thrombocythemia (ET), and primary myelofibrosis (PMF) whereas CALR and MPL mutants are found in ET and PMF. Other mutations in genes involved in epigenetic regulation, splicing, and signaling cooperate with the 3 MPN drivers and play a key role in the PMF pathogenesis. Mutations in epigenetic regulators TET2 and DNMT3A are involved in disease initiation and may precede the acquisition of JAK2V617F. Other mutations in epigenetic regulators such as EZH2 and ASXL1 also play a role in disease initiation and disease progression. Mutations in the splicing machinery are predominantly found in PMF and are implicated in the development of anemia or pancytopenia. Both heterogeneity of classical MPNs and prognosis are determined by a specific genomic landscape, that is, type of MPN driver mutations, association with other mutations, and their order of acquisition. However, factors other than somatic mutations play an important role in disease initiation as well as disease progression such as germ line predisposition, inflammation, and aging. Delineation of these environmental factors will be important to better understand the precise pathogenesis of MPN.
Introduction
The classical myeloproliferative neoplasms (MPNs), also called BCR-ABL− MPNs, are the most frequent diseases among the myeloproliferative disorders. MPNs are characterized by excessive production of terminally differentiated blood cells that are fully functional. Classical MPNs have been classified into 3 entities: polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), which have frequent disease-related complications, such as venous and arterial thrombosis, hemorrhages, and transformation to acute myeloid leukemia (AML).1 All MPN entities arise from a single somatically mutated hematopoietic stem cell (HSC) that clonally expands and gives rise to virtually all myeloid cells, and B and natural killer (NK) cells. The clonal expansion of the MPN HSC is accompanied by single or multilineage hyperplasia. PV is characterized not only by an excess of erythrocytes and predominant erythroid lineage involvement, but is also associated with a variable hyperplasia of the megakaryocytic/granulocytic lineages. ET is characterized by an increased platelet count with a megakaryocytic hyperplasia, whereas PMF is a more heterogeneous disorder both by its clinical and biological characteristics, defined by the presence of bone marrow fibrosis (excess of collagen fibers) and megakaryocytic hyperplasia. Myeloproliferation in PMF initially predominates in the bone marrow and later expands to extramedullary sites, such as the spleen. Although the clinical presentations of the 3 MPN entities are different in their typical forms, establishment of precise diagnosis at disease onset is often challenging. This is reflected by the 2016 revision of the World Health Organization (WHO) diagnostic criteria for MPN.2 In many cases, a continuum between these disease subtypes can be observed, as documented by the progression of ET and PV to secondary myelofibrosis (MF). Furthermore, boundaries between these 3 disorders cannot be well established, especially between ET and PMF. Thus, transitional entities may emerge describing disease states such as prefibrotic PMF (or early PMF) that displays an ET phenotype at diagnosis with typical bone marrow histology, but with a high probability of progression to MF and worse prognosis than true ET.3
Somatic mutations are responsible for clonal expansion of HSCs not only in MPNs, but also in most types of myeloid malignancies. High-resolution genome analysis using microarray and next-generation sequencing (NGS) resulted in the discovery of several gene mutations across all myeloid malignancies. Only relatively few of these gene mutations are found associated with a single or limited number of disease entities. Therefore, we classify these mutations into those that are “restricted to MPN” and those that are “nonrestricted” and are found also in other myeloid malignancies (Table 1).
Somatic mutations in MPN
| Gene function and symbol . | Location . | Type of mutations . | Protein function . | Frequency . | Consequences . | Reference . |
|---|---|---|---|---|---|---|
| Signaling MPN driver | ||||||
| JAK2 | 9p24 | JAK2V617F | Tyrosine kinase associated with cytokine receptors | 95% PV, 50%-60% PMF and ET | Increased RBC, platelet, and granulocyte production | 4,,-7 |
| JAK2 exon 12 | 3% PV | Increased RBC | 17 | |||
| MPL | 1p34 | MPL515L/K/A/R MPLS505N | TPOR | 2%-3% ET | Increased platelet production | 19, 22, 23 |
| Other missense mutations | 3-5% PMF | 25 | ||||
| CALR | 19p13 | Indel exon 9 | Mutant: activator of MPL | 20%-25% ET, 25%-30% PMF | Increased platelet production | 28, 29 |
| Other signaling | ||||||
| LNK | 12q24 | Missense (loss of function) deletion | Negative regulator of JAK2 | 1% ET, 2% PMF | Synergy with JAK2V617F-Disease progression | 35 |
| CBL | 11q23;3 | Missense (loss of function) | Cytokine receptor internalization | 4% PMF | Disease progression (progression to AML) | 110 |
| NRAS | 1p13.2 | Missense (activation) | ERK/MAPK signaling | Rare PMF | Progression to leukemia (5%-10% in secondary AML) | 65 |
| NF1 | 17q11 | Missense deletion | ERK/MAPK signaling | Rare PMF | Progression to leukemia (5%-10% in secondary AML) | 66 |
| FLT3 | 13q12 | FLT3-ITD | Cytokine receptor (FLT3-L) | MPN (<3%) | Progression to leukemia (10%-15% in secondary AML) | 100 |
| DNA methylation | ||||||
| TET2 | 4q24 | Missense, nonsense deletion | α-Ketoglutarate–dependent dioxygenase | 10%-20% MPN (ET, PV, and PMF) | Initiation, | 70 |
| Oxidation of 5mC into 5hmC and active 5mC demethylation | Mutations on 2 alleles associated with progression | |||||
| DNMT3A | 2p23 | Missense, hotspot | DNA methylase, de novo methylation | 5%-10% MPN (ET, PV, and PMF) | Initiation | 74 |
| IDH1 | 2q33.3 | Missense, hotspot | Neomorphic enzyme, generation of 2-hydroxyglutarate blocking α-ketoglutarate–dependent enzymes | 1%-3% PMF | Initiation, Disease progression | 111 |
| IDH2 | 15q26.1 | Missense, hotspot | Neomorphic enzyme, generation of 2-hydroxyglutarate blocking α-ketoglutarate–dependent enzymes | 1%-3% PMF | Initiation, Disease progression | 111 |
| Histone modifications | ||||||
| EZH2 | 7q35-36 | Missense, indel | H3K27 methyltransferase, loss of function | 5%-10% PMF | Initiation | 82, 83 |
| Disease progression | ||||||
| ASXL1 | 20q11 | Nonsense/ indel | Chromatin-binding protein associated with PRC1 and 2 | 25% PMF | Initiation | 69 |
| 1%-3% ET/PV | Rapid progression | |||||
| Transcription factors | ||||||
| TP53 | 17p13.1 | Missense/ indel | Transcription factor regulating cell cycle, DNA repair and apoptosis | <5% (20% of sAML) | Progression to leukemia (mutations on both alleles) complex karyotype | 98 |
| CUX1 | 7q22 | Deletion 7p | Transcription factor regulating TP53 and ATM | <3% | Progression to leukemia | 112 |
| IKZF1 | 7p12.2 | Deletion 7p, indel | Master transcription factor in lymphopoiesis | <3% | Progression to leukemia | 112 |
| ETV6 | 12p13 | Missense/ indel | Transcription factor of the ETs family | <3% | Progression to leukemia | 112 |
| RUNX1 | 21q22.3 | Nonsense/ missense/ indel | Master transcription factor in hematopoiesis | <3% (10% of sAML) | Progression to leukemia | 112 |
| RNA splicing | ||||||
| SRSF2 | 17q25.1 | Missense, hotspot | Serine/arginine-rich pre- RNA splicing factor | <2% ET 10%-15% PMF (association with IDH mutations) | Initiation? Progression | 94 |
| SF3B1 | 2q33.1 | Missense | RNA-splicing factor 3b subunit 1, part of U2 | <3% ET | Phenotypic change (anemia) | 113 |
| U2AF1 | 21q22.3 | Missense | U2 small nuclear RNA-splicing factor | 10%-15% PMF | Phenotypic change (anemia) | 94 |
| Gene function and symbol . | Location . | Type of mutations . | Protein function . | Frequency . | Consequences . | Reference . |
|---|---|---|---|---|---|---|
| Signaling MPN driver | ||||||
| JAK2 | 9p24 | JAK2V617F | Tyrosine kinase associated with cytokine receptors | 95% PV, 50%-60% PMF and ET | Increased RBC, platelet, and granulocyte production | 4,,-7 |
| JAK2 exon 12 | 3% PV | Increased RBC | 17 | |||
| MPL | 1p34 | MPL515L/K/A/R MPLS505N | TPOR | 2%-3% ET | Increased platelet production | 19, 22, 23 |
| Other missense mutations | 3-5% PMF | 25 | ||||
| CALR | 19p13 | Indel exon 9 | Mutant: activator of MPL | 20%-25% ET, 25%-30% PMF | Increased platelet production | 28, 29 |
| Other signaling | ||||||
| LNK | 12q24 | Missense (loss of function) deletion | Negative regulator of JAK2 | 1% ET, 2% PMF | Synergy with JAK2V617F-Disease progression | 35 |
| CBL | 11q23;3 | Missense (loss of function) | Cytokine receptor internalization | 4% PMF | Disease progression (progression to AML) | 110 |
| NRAS | 1p13.2 | Missense (activation) | ERK/MAPK signaling | Rare PMF | Progression to leukemia (5%-10% in secondary AML) | 65 |
| NF1 | 17q11 | Missense deletion | ERK/MAPK signaling | Rare PMF | Progression to leukemia (5%-10% in secondary AML) | 66 |
| FLT3 | 13q12 | FLT3-ITD | Cytokine receptor (FLT3-L) | MPN (<3%) | Progression to leukemia (10%-15% in secondary AML) | 100 |
| DNA methylation | ||||||
| TET2 | 4q24 | Missense, nonsense deletion | α-Ketoglutarate–dependent dioxygenase | 10%-20% MPN (ET, PV, and PMF) | Initiation, | 70 |
| Oxidation of 5mC into 5hmC and active 5mC demethylation | Mutations on 2 alleles associated with progression | |||||
| DNMT3A | 2p23 | Missense, hotspot | DNA methylase, de novo methylation | 5%-10% MPN (ET, PV, and PMF) | Initiation | 74 |
| IDH1 | 2q33.3 | Missense, hotspot | Neomorphic enzyme, generation of 2-hydroxyglutarate blocking α-ketoglutarate–dependent enzymes | 1%-3% PMF | Initiation, Disease progression | 111 |
| IDH2 | 15q26.1 | Missense, hotspot | Neomorphic enzyme, generation of 2-hydroxyglutarate blocking α-ketoglutarate–dependent enzymes | 1%-3% PMF | Initiation, Disease progression | 111 |
| Histone modifications | ||||||
| EZH2 | 7q35-36 | Missense, indel | H3K27 methyltransferase, loss of function | 5%-10% PMF | Initiation | 82, 83 |
| Disease progression | ||||||
| ASXL1 | 20q11 | Nonsense/ indel | Chromatin-binding protein associated with PRC1 and 2 | 25% PMF | Initiation | 69 |
| 1%-3% ET/PV | Rapid progression | |||||
| Transcription factors | ||||||
| TP53 | 17p13.1 | Missense/ indel | Transcription factor regulating cell cycle, DNA repair and apoptosis | <5% (20% of sAML) | Progression to leukemia (mutations on both alleles) complex karyotype | 98 |
| CUX1 | 7q22 | Deletion 7p | Transcription factor regulating TP53 and ATM | <3% | Progression to leukemia | 112 |
| IKZF1 | 7p12.2 | Deletion 7p, indel | Master transcription factor in lymphopoiesis | <3% | Progression to leukemia | 112 |
| ETV6 | 12p13 | Missense/ indel | Transcription factor of the ETs family | <3% | Progression to leukemia | 112 |
| RUNX1 | 21q22.3 | Nonsense/ missense/ indel | Master transcription factor in hematopoiesis | <3% (10% of sAML) | Progression to leukemia | 112 |
| RNA splicing | ||||||
| SRSF2 | 17q25.1 | Missense, hotspot | Serine/arginine-rich pre- RNA splicing factor | <2% ET 10%-15% PMF (association with IDH mutations) | Initiation? Progression | 94 |
| SF3B1 | 2q33.1 | Missense | RNA-splicing factor 3b subunit 1, part of U2 | <3% ET | Phenotypic change (anemia) | 113 |
| U2AF1 | 21q22.3 | Missense | U2 small nuclear RNA-splicing factor | 10%-15% PMF | Phenotypic change (anemia) | 94 |
ATM, ataxia-telangiectasia mutated; RBC, red blood cell; sAML, secondary AML.
Mutations restricted to MPNs and their role in the MPN pathogenesis
JAK2 gene mutations
Before 2005, the molecular pathogenesis of the BCR-ABL1− classical MPNs was unknown. In 2005, a major breakthrough was the discovery of a G to T somatic mutation at nucleotide 1849, in exon 14 of JAK2, resulting in the substitution of valine to phenylalanine at codon 617 (JAK2V617F) in the pseudokinase domain.4-7 This mutation can be found in around 70% of MPNs: 95% of PV and 50% to 60% of ET and PMF. The JAK2V617F mutation often undergoes a transition from heterozygosity to homozygosity due to occurrence of mitotic recombination resulting in copy-neutral loss of heterozygosity along a variable size region on the short arm of chromosome 9 (9pLOH).5,8 The JAK2V617F mutation arises in a multipotent hematopoietic progenitor, is present in all myeloid lineages, and can be also detected in lymphoid cells, mainly B and NK cells and more rarely and later in disease in T cells. It is absent in nonhematopoietic cells although it has been found in endothelial cells of the spleen and of splanchnic veins of patients with MF and/or splanchnic thrombosis as the Budd-Chiari syndrome.9 The variant allele frequency (VAF) in granulocytes is highly variable, with a continuum between the threshold of detection (around 1%) to 100%. The VAF is usually low in ET (around 25%), is higher in PV (frequently over 50%), and is close to 100% in post-PV or post-ET MF. In PMF, the VAF is variable, but frequently high.10 The JAK2V617F is mainly restricted to classical MPNs with the exception of refractory anemia with ring sideroblasts and thrombocytosis (RARS-T).11 It can rarely be found in some other malignant hemopathies.12 However, JAK2V617F has been detected at very low level (lower than 1%) in the normal population, including in a neonate.13-15 It is 1 of the most frequent mutations found in the clonal hematopoiesis associated with aging (clonal hematopoiesis of indeterminate potential).16
JAK2 exon 12 mutations have been also found in MPNs and are present in the majority of JAK2V617F− PV.17 They are not usually associated with ET and PMF, but JAK2 exon 12 mutation can progress to secondary MF.18 Several mutations have been described, most of them being in-frame small insertions and deletions. Exon 12 JAK2 mutations are all located in the linker between the Src homology 2 (SH2) and the pseudokinase domains, a region between amino acids 536 and 547, around Lys 539. The most frequent mutations being the N542-E543del (23%), E543-D544del (11%), and F537-K539delinsL and K539L (10%), respectively. The VAF in granulocytes can be low and their detection may require specific techniques.
Thrombopoietin receptor gene (MPL) mutations
Two main types of myeloproliferative leukemia virus (MPL; the thrombopoietin [TPO] receptor [TPOR]) mutations located in exon 10 have been associated with MPNs. The most frequent are mutations on the tryptophan W515 located at the boundary of the transmembrane and the cytosolic domains of MPL, the most prominent mutations being MPLW515L and K.19 However, several other substitutions have been described such as W515R, W515A, and W515G.20 Mutations are usually heterozygous, but can be homozygous during disease progression.21 Mutations on MPLW515 are restricted to ET (around 3%) and PMF (around 5%) and can be also found in RARS-T.22-24 The other mutation, MPLS505N, is even rarer and is located in the transmembrane domain, stabilizing receptors in active dimeric orientations. Originally, this mutation was described in hereditary form of thrombocytosis as a germ line mutation.25 Later on, acquired MPLS505N somatic mutations were described in ET in <1% of the cases.22 ,23 This underscores the similarities in mechanisms of thrombocytosis between hereditary thrombocytosis and ET. More recently, a number of “noncanonical” mutations of MPL were found in triple-negative ET (ET negative for JAK2V617F, calreticulin [CALR] mutations, and MPLW515L/K / MPLS505N). These mutations are rare and lead to amino acid changes either in the extracellular (S204 or E230) or in the intracellular (Y591) domains.26,27 These noncanonical mutations are more often acquired and lead to a true MPN. In addition, germ line noncanonical mutations of MPL can also be detected, strongly suggesting that these triple-negative ET cases are in fact hereditary thrombocytosis.
Mutations in the calreticulin gene: CALR
At the end of 2013, frameshift mutations in the CALR gene were discovered in the majority of JAK2− and MPL− ET and PMF (50%-60% ET and 75% PMF).28,29 There are >50 mutations now described, but all are located in exon 9 inducing a +1 (−1+2) frameshift. At the time of writing, only the mutations leading to this +1 frameshift are known to be pathogenic. The other nonpathogenic mutations are usually germ line variants of CALR. Specific shifting of the reading frame leads to a new C terminus devoid of the KDEL motif, important for the protein retention in the endoplasmic reticulum (ER). The mutated CALR protein contains a common new amino acid sequence that bears positive charges. In contrast to wild-type (WT) CALR, the negative charges required for calcium binding are lost at different extents, depending on the mutant. The 2 most frequent mutations correspond to a 52-bp deletion (p.L367fs*46), also called type 1 and a 5-bp insertion (p.K385fs*47), also called type 2. They represent nearly the 2 extremes of modifications observed in exon 9: the del52 having lost most of the WT exon 9 sequence and calcium-binding sites, and ins5 being closer to the WT sequence having kept around 50% of negative charges. According to these structural changes, the other mutations have been classified as type 1–like and type 2–like using algorithms based on the preservation of an α helix close to WT in type 2–like mutation.30 There are great differences in the frequency between type 1 and type 2 mutations in ET and PMF: in ET, type 1 and type 2 mutations are closely distributed (55% vs 35%), whereas in PMF, type 1 are largely predominant (75% vs 15%).31 The presence of CALR mutants in exceptional cases of PV has been reported, but their role in the pathogenesis of the PV remains unclear.32
CALR mutations are usually heterozygous although a few cases of homozygous mutations have been observed, more particularly for type 2 mutations.28,29 Interestingly, homozygous CALR mutations are associated with a defect in myeloperoxidase accumulation in primary granules of granulocytes, and thus, can be suspected in an ET when a “myeloperoxidase deficiency” is detected by using a flow cytometer–based hematology analyzer.33
In contrast to JAK2V617F, the VAF for mutant CALR in ET is high, on average around 40%.10 In rare cases of ET, the VAF can be lower, but almost never below 15%. This implies that CALR mutants might give a stronger clonal advantage when compared with JAK2V617F. The molecular diagnosis is rendered easier by this high VAF and can be performed by numerous techniques and routinely by sizing fluorescently labeled polymerase chain reaction products of exon 9 of CALR.28 A few CALR polymorphisms have been detected not only as single-nucleotide polymorphisms, but also in phase deletions of exon 9.34 There is presently no evidence that these variants have any role in pathogenesis.
LNK mutations
In 2010, somatic mutations in exon 2 of LNK (SH2B3), an adaptor protein which regulates JAK2 activation, were detected in 2 patients (PMF and ET).35 These mutations had a functional impact on proliferation and were considered as the driver of the disease. Recent studies have questioned the role of LNK as a true driver of the disease.36 LNK mutations have been found in many different types of hemopathies, more particularly in erythrocytosis.37 Some of these mutations can be germ line, including the initially described LNKE208Q; some others are secondary somatic mutations associated with JAK2V617F or CALR mutations, more particularly during disease progression.38 Thus, rather than primary drivers, LNK mutations appear more as predisposition mutations when germ line or secondary mutations, increasing the pathogenicity of JAK2V617F and CALR when acquired.36
MPN-restricted mutations activate JAK2 kinase–dependent cytokine receptor pathways
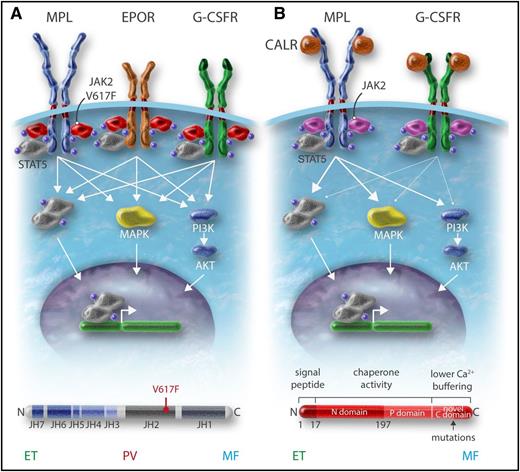
JAK2 is a member of the JAK family characterized by 2 kinase domains: 1 catalytically active at the C terminus and 1 catalytically inactive (or very weak) pseudokinase that prevents self-activation of the kinase domain.39 At the N terminus, JAKs possess a four-point-one ezrin radixin moesin (FERM)-like domain and a SH2-like domain. The noncovalent attachment of JAKs to cytokine receptors depends on the FERM domain. The JAK family kinases can be considered as the catalytic part of the hematopoietic cytokine receptor family as they are constitutively bound to the receptors intracellularly. In addition, the association of JAKs with receptors is important for their proper trafficking to the cell surface. Homodimeric receptors such as erythropoietin (EPO) receptor (EPOR), MPL, and granulocyte colony-stimulating factor receptor (G-CSFR) use JAK2, whereas heteromeric receptors use JAK1 and JAK2/tyrosine kinase 2 (TYK2) or JAK3. Cytokine binding induces changes in the conformation of receptors, which activate the JAKs bound to the receptor by trans-phosphorylation. The activated JAKs will phosphorylate the receptors, which are subsequently used as a docking site for other signaling molecules, more particularly the STATs. In the canonical pathway, STATs are phosphorylated by the JAKs, which induce their homodimerization or heterodimerization and subsequent trafficking to the nucleus where they regulate transcription of their target genes (Figure 1). The pseudokinase domain of JAK2 has 2 roles: 1 is to inhibit the kinase domain and the other is to promote cytokine-dependent activation. V617F activates JAK2 by a mechanism, which is not completely understood, but both structural and functional data have established that the first conformational change induced by V617F involves the helix C of the pseudokinase domain.40,41 Mutations in exon 12 are located upstream of the pseudokinase domain, and the mechanism of activation appears different than that for JAK2V617F.42 JAK2V617F or JAK2 exon 12 mutations, when expressed in interleukin-3 (IL-3)-dependent cell lines, induce cytokine hypersensitivity or cytokine independence. The presence of homodimeric receptors such as EPOR greatly facilitates this biological effect.43 The presence of a cytokine receptor is absolutely required to induce signaling by JAK2V617F at low expression levels. JAK2V617F induces constitutive activation of STATs, and of the phosphatidylinositol 3-kinase (PI3K) and MAPK pathways. This cytokine hypersensitivity or independence induced by JAK2V617F in cell lines mimics the EPO hypersensitivity or independence previously described in PV erythroid progenitors.4,44 In addition to this activation of the canonical pathway of cytokine receptors, 2 reports have described that JAK2V617F acts on chromatin by phosphorylating H3Y41, decreasing the binding of heterochromatin protein 1a and PRMT5, impairing its ability to methylate histone substrates.45,46 It is unknown whether these effects on chromatin are really important in the pathogenesis of MPNs.
Role of cytokine receptors in the oncogenic properties of JAK2V617F and CALR mutants. (A) JAK2V617F activates signaling through the 3 main homodimeric receptors EPOR, MPL, and G-CSFR, which are involved in erythrocytosis, thrombocytosis, and neutrophilia, respectively. (B) The CALR mutants mainly activate MPL and at a low level the G-CSFR but not the EPOR, explaining the thrombocytosis associated with these mutants. Professional illustration by Somersault18:24.
Role of cytokine receptors in the oncogenic properties of JAK2V617F and CALR mutants. (A) JAK2V617F activates signaling through the 3 main homodimeric receptors EPOR, MPL, and G-CSFR, which are involved in erythrocytosis, thrombocytosis, and neutrophilia, respectively. (B) The CALR mutants mainly activate MPL and at a low level the G-CSFR but not the EPOR, explaining the thrombocytosis associated with these mutants. Professional illustration by Somersault18:24.
Activation of MPL by TPO induces the activation of JAK2 and TYK2, which are constitutively bound to the receptor, but only the activation of JAK2 is indispensable for signaling and induction of proliferation by MPL. MPLW515 is located in the amphipathic domain of MPL, which prevents spontaneous activation of MPL.47 All substitutions of W515, except W515C and W515P, will lead to the activation of MPL in the absence of TPO and will activate JAK2. The S505N mutation induces a stable active dimer. However, both types of mutations activate constitutive signaling through induction of productive dimerization of the transmembrane helix of MPL.20,25
CALR in contrast is not a signaling molecule, but an ER chaperone involved in the quality control of N-glycosylated proteins and in calcium storage. The initial publication on CALR mutations has shown that CALRdel52 was able to activate STAT5 and this was impeded by JAK2 inhibition.28 Recent studies have established that CALR mutants can activate MPL and subsequently JAK248-52 (Figure 2). Mutant CALR requires both its mutant C terminus and MPL for oncogenic transformation.49,50,53 WT CALR and mutant CALR bind to the N-glycosylated residue of the extracellular domain of MPL in the ER, and the lectin-binding activity of CALR mutants as well as the new tail are required for MPL activation49 (Figure 2). This binding is reinforced by the new C terminus and its positive charges, which lead to activation of the receptor.50,53 It is unknown at which cellular compartment MPL is activated, but there is evidence that CALR mutants behave as an abnormal chaperone and traffics with MPL to the cell surface.47 In this case, MPL activation can occur anywhere from the ER to the cell surface. Moreover, CALR mutants are secreted and may induce cytokine secretion by monocytes.54 CALR mutants also appear to slightly activate the G-CSFR, but not the other cytokine receptors.49 Whether this activation of G-CSFR plays a role in the pathogenesis of mutant CALR+ MPN is unknown (Figure 1).
The CALR mutants bind and activate MPL. (A) WT CALR binds to MPL in the ER by interaction of its lectin domain with MPL N-glycosylation, controls the quality of the proteins, and then comes away from the receptor, which traffics to the cell surface mainly by the conventional route to the cell surface. A completely glycosylated MPL (mature form) is expressed at the cell surface where it binds TPO to be activated. CALR mutants bind to MPL in the ER by the same interaction as the WT but the interaction is reinforced by the new C terminus. (B) The mutant CALR remains bound to MPL and probably traffics to the membrane both conventionally and unconventionally (autophagosomes and pre-Golgi) to the membrane. This leads to the presence of an immature MPL form on which the mutant CALR is bound. Activation of MPL is dependent on the new C terminus and also takes place on the membrane (Stefan Constantinescu, W.V., unpublished results). In cell lines, this leads to activation of mainly the STAT pathway whereas the extracellular signal-regulated kinase (ERK) is slightly activated and the PI3K pathway even less. In cell lines, MPL that binds mutant CALR can only be slightly activated by TPO. Professional illustration by Somersault18:24.
The CALR mutants bind and activate MPL. (A) WT CALR binds to MPL in the ER by interaction of its lectin domain with MPL N-glycosylation, controls the quality of the proteins, and then comes away from the receptor, which traffics to the cell surface mainly by the conventional route to the cell surface. A completely glycosylated MPL (mature form) is expressed at the cell surface where it binds TPO to be activated. CALR mutants bind to MPL in the ER by the same interaction as the WT but the interaction is reinforced by the new C terminus. (B) The mutant CALR remains bound to MPL and probably traffics to the membrane both conventionally and unconventionally (autophagosomes and pre-Golgi) to the membrane. This leads to the presence of an immature MPL form on which the mutant CALR is bound. Activation of MPL is dependent on the new C terminus and also takes place on the membrane (Stefan Constantinescu, W.V., unpublished results). In cell lines, this leads to activation of mainly the STAT pathway whereas the extracellular signal-regulated kinase (ERK) is slightly activated and the PI3K pathway even less. In cell lines, MPL that binds mutant CALR can only be slightly activated by TPO. Professional illustration by Somersault18:24.
Overall, the main MPN-associated mutations, JAK2V617F, JAK2 exon 12 mutants, MPLW515L/K, and CALR mutants, activate the cytokine/receptor/JAK2 pathways and their downstream signaling, JAK2V617F, through the 3 homodimeric receptors (EPOR, MPL, G-CSFR) and CALR mutants through mainly MPL (Figure 1). This may explain also why these different mutations are usually mutually exclusive. However, rarely, 2 of these mutations can be detected in the same patient, but usually they are present in different clones or subclones.
MPN-restricted mutations drive myeloproliferative disorders and give rise to different diseases in murine models
All 3 MPN mutation types have been expressed in mice to investigate whether they are drivers of MPNs and to model the disease (Table 2). JAK2V617F has been expressed in mouse hematopoiesis by several approaches: retroviral transduction of HSC followed by bone marrow transplantation, transgenic mice, and constitutive or inducible knock-in (KI) mice. Whatever the model, expression of JAK2V617F induces a myeloproliferative phenotype with certain differences. The most frequent phenotype has been a PV-like disorder progressing to MF.55,56 A correlation between the level of expression and the phenotype has been found: low expression being associated with an ET-like phenotype and a higher expression (30% of the WT or more) with a PV-like phenotype.57 Further phenotypic variability may be related to the use of human vs murine JAK2V617F. The heterozygous KI of the human JAK2V61F displays a very mild ET phenotype and the homozygous KI induces an erythrocytosis.58 Thus, in mouse models, JAK2V617F is the driver of the myeloproliferative disorder and the level of expression in great part determines the phenotype of the MPN, a situation close to the human setting. Recently, JAK2 exon 12 transgenic mice have been obtained that developed isolated erythrocytosis.59 The erythroid phenotype was even more marked than in JAK2V617F transgenic mice. This was attributed to important changes in iron metabolism including decreased expression in hepcidin and increased expression in transferrin receptor-1 and erythroferrone that favor iron availability for erythroblasts.
Mouse models
| Mouse models . | Phenotype . | Reference . |
|---|---|---|
| JAK2V617F | ||
| Retrovirus | ||
| C57Bl/6 | PV progressing to MF | 114,-116 |
| BALB/c | Severe lethal PV to MF | 115, 116 |
| Transgenic | ||
| Vav huJAK2V617F | ET/PV progressing to MF | 117 |
| H2K muJak2V617F | ET/PV progressing to MF | 118 |
| Inducible JAK2min promoter huJAK2V617F | ||
| Vav Cre | ET | 57 |
| Mx Cre | PV with thrombocytosis | 57 |
| Pf4Cre (MK specific) | No phenotype | 119 |
| Mild ET and a PV after transplantation | 120 | |
| Tie2Cre (HSC and endothelial cell) | ET with neutrophilia progressing to osteopetrosis with bleeding diathesis | 119 |
| Knock-in | ||
| Constitutive muJak2V617F | PV with thrombocytosis progressing to MF | 121 |
| Germ line induction of muJAK2V617F | PV progressing to MF | 55 |
| E2Acre | ||
| Germ line induction of huJAK2V617F (cDNA) | ||
| Stella Cre | ||
| Heterozygous | Mild ET | 122 |
| Homozygous | PV | 122 |
| muJAK2V617F | ||
| Mx Cre | ||
| Heterozygous | ET/PV progressing to MF | 123 |
| Homozygous | Severe PV progressing to MF | 123 |
| EpoR Cre (erythroid specific) | ||
| Heterozygous | Mild erythrocytosis | 124 |
| Homozygous | Erythrocytosis | 124 |
| muJAK2V617F heterozygous | ||
| Vav Cre | ET progressing to PV progressing to MF | 56 |
| Scl Cre | PV progressing to MF | 125 |
| huJAK2V617F (cDNA) heterozygous | Mild ET progressing in PV (10% cases) | 58 |
| Mx Cre | ||
| CALR | ||
| Retrovirus | ||
| CALRdel52 | Mild ET, ET progressing to MF | 50,51 |
| CALRins5 | Mild ET | 51 |
| Transgenic | ||
| H-2Kb promoter | ET | 126 |
| Knock-in | ||
| Constitutive muCALRdel 61 | ET | 60 |
| Inducible humanized CALRdel52 | Caroline Marty, Isabelle Plo, Harini Nivarthi, R.K., W.V., unpublished results | |
| Vav Cre | ET (short-term follow-up) | |
| SCL CreER | ET (short-term follow-up |
| Mouse models . | Phenotype . | Reference . |
|---|---|---|
| JAK2V617F | ||
| Retrovirus | ||
| C57Bl/6 | PV progressing to MF | 114,-116 |
| BALB/c | Severe lethal PV to MF | 115, 116 |
| Transgenic | ||
| Vav huJAK2V617F | ET/PV progressing to MF | 117 |
| H2K muJak2V617F | ET/PV progressing to MF | 118 |
| Inducible JAK2min promoter huJAK2V617F | ||
| Vav Cre | ET | 57 |
| Mx Cre | PV with thrombocytosis | 57 |
| Pf4Cre (MK specific) | No phenotype | 119 |
| Mild ET and a PV after transplantation | 120 | |
| Tie2Cre (HSC and endothelial cell) | ET with neutrophilia progressing to osteopetrosis with bleeding diathesis | 119 |
| Knock-in | ||
| Constitutive muJak2V617F | PV with thrombocytosis progressing to MF | 121 |
| Germ line induction of muJAK2V617F | PV progressing to MF | 55 |
| E2Acre | ||
| Germ line induction of huJAK2V617F (cDNA) | ||
| Stella Cre | ||
| Heterozygous | Mild ET | 122 |
| Homozygous | PV | 122 |
| muJAK2V617F | ||
| Mx Cre | ||
| Heterozygous | ET/PV progressing to MF | 123 |
| Homozygous | Severe PV progressing to MF | 123 |
| EpoR Cre (erythroid specific) | ||
| Heterozygous | Mild erythrocytosis | 124 |
| Homozygous | Erythrocytosis | 124 |
| muJAK2V617F heterozygous | ||
| Vav Cre | ET progressing to PV progressing to MF | 56 |
| Scl Cre | PV progressing to MF | 125 |
| huJAK2V617F (cDNA) heterozygous | Mild ET progressing in PV (10% cases) | 58 |
| Mx Cre | ||
| CALR | ||
| Retrovirus | ||
| CALRdel52 | Mild ET, ET progressing to MF | 50,51 |
| CALRins5 | Mild ET | 51 |
| Transgenic | ||
| H-2Kb promoter | ET | 126 |
| Knock-in | ||
| Constitutive muCALRdel 61 | ET | 60 |
| Inducible humanized CALRdel52 | Caroline Marty, Isabelle Plo, Harini Nivarthi, R.K., W.V., unpublished results | |
| Vav Cre | ET (short-term follow-up) | |
| SCL CreER | ET (short-term follow-up |
cDNA, complementary DNA.
For MPLW515K/L/A, only retroviral models have been described. Although these models are far from ideal because they lead to the expression of MPLW515 mutant in all types of hematopoietic cells, an ET-like disorder rapidly progressing to MF is usually observed, a phenotype close to the mice expressing a high level of TPO.19
For CALR mutants, retroviral models have been used to induce a myeloproliferative disorder reminiscent of an ET-like phenotype, which in the case of CALRdel52 may progress to MF. In this mouse model, the presence of MPL is indispensable for disease development.50,51 A recent transgenic model for CALRdel52 also led to an ET-like phenotype that was ameliorated by ruxolitinib, similar results being obtained in del61 or del52 KI mice60 (Caroline Marty, Isabelle Plo, Harini Nivarthi, W.V., R.K., unpublished results).
In conclusion, the 3 MPN oncogenes are true drivers of the disease phenotype with JAK2 exon 12 giving only an erythrocytosis phenotype, JAK2V617F giving rise to ET, PV, and MF, whereas CALR mutant and MPLW515L/K/A are associated with ET and MF, resembling the phenotype observed in patients. The current data sustain the hypothesis that the phenotype of MPNs is mostly related to the types of receptors that are activated. However, in all models, MPL activation plays a central role by its role on HSCs and on megakaryocytes (MKs).19,51,61 MK hyperplasia plays a central role in the pathogenesis of MPNs not only by increasing platelet production, but also by serving as a niche for HSCs, by remodeling bone marrow leading to MF and also by inducing neo-osteogenesis (Figure 3). However, in none of the models did mice develop a PMF, whereas they did develop secondary MF (post-ET or post-PV); no leukemic transformation is observed during disease progression. The different STATs may play an important role in disease phenotype: STAT5 being required for the PV development, STAT3 inhibiting the thrombocytosis, and STAT1 increasing the thrombocytosis.62,63
MKs play a central role in MPN pathogenesis. The MPL/JAK2 pathway is activated by the 3 MPN-restricted mutations (JAK2V617F, CALR mutants, and MPL mutants) placing MK hyperplasia and eventually dysplasia as a central determinant in MPN. MKs are mainly involved in platelet production, and also play an important role in the hematopoietic niche by regulating HSCs, remodeling the marrow by secretion of TGF-β1 and other cytokines (platelet-derived growth factor [PDGF], vascular endothelial growth factor [VEGF]) which ultimately lead to MF, and also inducing a neo-osteogenesis by inducing an osteoblastic differentiation through TGF-β1 and inhibiting osteoclast differentiation through osteoprotegerin. Furthermore, MKs secrete numerous inflammatory cytokines such as IL1α. The mechanisms of local activation of TGF-β1 are poorly known. OPG, osteoprotegerin; RANK, receptor activator of NF-κB; RANKL, RANK ligand; TGF, transforming growth factor; TSP, thrombospondin. Professional illustration by Somersault18:24.
MKs play a central role in MPN pathogenesis. The MPL/JAK2 pathway is activated by the 3 MPN-restricted mutations (JAK2V617F, CALR mutants, and MPL mutants) placing MK hyperplasia and eventually dysplasia as a central determinant in MPN. MKs are mainly involved in platelet production, and also play an important role in the hematopoietic niche by regulating HSCs, remodeling the marrow by secretion of TGF-β1 and other cytokines (platelet-derived growth factor [PDGF], vascular endothelial growth factor [VEGF]) which ultimately lead to MF, and also inducing a neo-osteogenesis by inducing an osteoblastic differentiation through TGF-β1 and inhibiting osteoclast differentiation through osteoprotegerin. Furthermore, MKs secrete numerous inflammatory cytokines such as IL1α. The mechanisms of local activation of TGF-β1 are poorly known. OPG, osteoprotegerin; RANK, receptor activator of NF-κB; RANKL, RANK ligand; TGF, transforming growth factor; TSP, thrombospondin. Professional illustration by Somersault18:24.
“Nonrestricted” acquired mutations and their role in the MPN pathogenesis
The 3 main driver mutations do not explain the entire heterogeneity of the classical BCR-ABL− MPNs. The development of NGS, as well as other whole-genome analysis techniques, allowed the identification of several acquired mutations in MPNs (Table 1). However, none of these mutations is restricted to MPNs and they are even more frequent in myelodysplastic syndromes (MDSs) and AML. This underscores the continuum between the different myeloid malignancies, and most of these mutations are involved in phenotypic changes and disease progression in MPNs.
Mutations identified in myeloid malignancies target DNA methylation regulators (TET2, DNMT3A, IDH1/2), histone modifiers (Polycomb repressor complex 1 and 2 members, IDH1/2), transcription factors (TP53, CUX1, IKZF1, FOXP1, ETV6, RUNX1), proteins involved in signaling (NF1, NRAS, KRAS, LNK, CBL, FLT3), and splicing factors (SF3B1, SRSF2, U2AF1, ZRSR2) (Figure 4).64-66 As the majority of the mutations are loss of function and act as dominant-negative or via haploinsufficiency or complete homozygous loss, it seems that most of the mutated genes are myeloid tumor suppressors. In MPNs, targeted gene panel sequencing studies identified patients with multiple gene mutations. Although recurrent single-gene mutations were identified, individual gene mutations had too low frequency to allow convincing evidence of association with disease progression. However, the number of detected mutations (as an indirect measure of genetic complexity or progression of clonal evolution) allowed the identification of high-risk patients.67
Genes involved in epigenetic regulation and leukemic transformation. The mechanisms by which the genes involved in the epigenetic regulation described in Table 1 lead to modifications in gene regulation are detailed. Some genes involved in leukemic transformation (N-Ras pathway and transcription factors such as p53, RUNX1) are also described. MEK, MAPK/ERK-kinase; RAF, rapidly accelerated fibrosarcoma; SOS, Son of Sevenless; TF, transcription factor. Professional illustration by Somersault18:24.
Genes involved in epigenetic regulation and leukemic transformation. The mechanisms by which the genes involved in the epigenetic regulation described in Table 1 lead to modifications in gene regulation are detailed. Some genes involved in leukemic transformation (N-Ras pathway and transcription factors such as p53, RUNX1) are also described. MEK, MAPK/ERK-kinase; RAF, rapidly accelerated fibrosarcoma; SOS, Son of Sevenless; TF, transcription factor. Professional illustration by Somersault18:24.
However, in ET and PV, in the majority of cases, only 1 mutation in a MPN driver gene is found. In contrast, in the majority of PMF, more somatic mutations are found (3 mutations or more). Some differences are observed in the coexpression of mutations along the type of MPNs. An example obtained by whole-exome sequencing is illustrated in Table 3. However, in several studies using targeted gene panel sequencing studies, mutations in spliceosome genes appear very rare in PV, in comparison with ET. This may be due to the continuum between ET and RARS-T or prefibrosis/PMF. Similarly, in PMF, there are no major differences with respect to the association of JAK2V617F, and CALR or MPL mutants with the different mutations in epigenetic regulators. On the other hand, spliceosome genes mutations are extremely rare in CALR PMF in comparison with JAK2V617F. Thus, PMF appears not to be a pure MPN, but a mixed myeloproliferative/myelodysplastic syndrome. The presence of mutations in genes other than the 3 MPN-restricted driver genes increases the myelodysplastic features of the disease and the severity of the disease, explaining the continuity between MPN, MPN/MDS, and MDS (Figure 5).
Co-occurrence of mutations
| . | PV JAK2V617F . | ET JAK2V617F . | ET CALRmut . | ET MPLm . | PMF JAK2V617F . | PMF CALRmut . | PMF MPLm . |
|---|---|---|---|---|---|---|---|
| % (No. of patients) . | |||||||
| TET2 | 8.33 (4) | 5.26 (3) | 0 (0) | 0 (0) | 28.95 (11) | 2.63 (1) | 2.63 (1) |
| DNMT3A | 6.25 (3) | 7.02 (4) | 0 (0) | 3.51 (2) | 7.89 (3) | 0 (0) | 0 (0) |
| ASXL1 | 0 (0) | 3.51 (2) | 0 (0) | 1.75 (1) | 15.79 (6) | 7.89 (3) | 0 (0) |
| IDH1/IDH2 | 2.08 (1) | 1.75 (1) | 0 (0) | 0 (0) | 2.63 (1) | 0 (0) | 0 (0) |
| EZH2 | 2.08 (1) | 0 (0) | 1.75 (1) | 0 (0) | 2.63 (1) | 2.63 (1) | 0 (0) |
| SF3B1 | 4.17 (2) | 0 (0) | 0 (0) | 0 (0) | 2.63 (1) | 0 (0) | 0 (0) |
| U2AF1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7.89 (3) | 0 (0) | 0 (0) |
| . | PV JAK2V617F . | ET JAK2V617F . | ET CALRmut . | ET MPLm . | PMF JAK2V617F . | PMF CALRmut . | PMF MPLm . |
|---|---|---|---|---|---|---|---|
| % (No. of patients) . | |||||||
| TET2 | 8.33 (4) | 5.26 (3) | 0 (0) | 0 (0) | 28.95 (11) | 2.63 (1) | 2.63 (1) |
| DNMT3A | 6.25 (3) | 7.02 (4) | 0 (0) | 3.51 (2) | 7.89 (3) | 0 (0) | 0 (0) |
| ASXL1 | 0 (0) | 3.51 (2) | 0 (0) | 1.75 (1) | 15.79 (6) | 7.89 (3) | 0 (0) |
| IDH1/IDH2 | 2.08 (1) | 1.75 (1) | 0 (0) | 0 (0) | 2.63 (1) | 0 (0) | 0 (0) |
| EZH2 | 2.08 (1) | 0 (0) | 1.75 (1) | 0 (0) | 2.63 (1) | 2.63 (1) | 0 (0) |
| SF3B1 | 4.17 (2) | 0 (0) | 0 (0) | 0 (0) | 2.63 (1) | 0 (0) | 0 (0) |
| U2AF1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7.89 (3) | 0 (0) | 0 (0) |
Results are extracted from Nangalia et al29 because it is one of the rare studies in which all types of MPNs are available. Overall, 143 patients: PV, 43; ET, 57; and PMF, 38. Percentages are expressed for each MPN, irrespective of the MPN drive mutation. Thus, percentages are calculated on a very low number of patients due to the heterogeneity of the disorder, explaining differences with other studies, more particularly for the mutations in splicing genes.
Presence of myeloproliferative and myelodysplastic features in MPN. (A-C) The 3 MPN-restricted drivers lead to a myeloproliferative phenotype. All of the additional mutations in genes involved in epigenetics and splicing modify the differentiation and give a myelodysplastic phenotype even if some of them such as TET2, DNMT3A, and EZH2 clearly play an important role in initiation. Moreover, there are additional mutations involved in splicing that induce myelodysplastic features leading to MF, cytopenia, and eventually progression to leukemia. Professional illustration by Somersault18:24.
Presence of myeloproliferative and myelodysplastic features in MPN. (A-C) The 3 MPN-restricted drivers lead to a myeloproliferative phenotype. All of the additional mutations in genes involved in epigenetics and splicing modify the differentiation and give a myelodysplastic phenotype even if some of them such as TET2, DNMT3A, and EZH2 clearly play an important role in initiation. Moreover, there are additional mutations involved in splicing that induce myelodysplastic features leading to MF, cytopenia, and eventually progression to leukemia. Professional illustration by Somersault18:24.
It is beyond the scope of this review to cover all the recurrent gene mutations in detail. Therefore, we will focus on TET2 and DNMT3A, as they may represent the most important modifiers of the MPN-restricted oncogenic mutations, on EZH2 whose role has been recently studied in detail, and on addition of sex combs like 1 (ASXL1) and SRSF2, which are all associated with a poor prognosis in MF.
TET2 and DNMT3A mutations and their role in disease initiation
The TET and DNMT3A protein families play a central role in the regulation of DNA methylation at cytosine guanine dinucleotide (CpG) sequences (Figure 4): the DNMT3 protein family is involved in the methylation of cytosines (5mC), whereas the TET protein family in active demethylation through oxidation of 5mC into 5hmC.68 TET2 mutations have been the first mutations, along with ASXL1, to be identified in JAK2V617F+ MPNs.69,70 All TET2 mutations are loss-of-function point mutations or deletions, usually on 1 allele, but more rarely on both somatic alleles. TET2 mutations have been identified initially as mutations preceding JAK2V617F and are responsible for clonal dominance.70 Their frequency ranges from 10% to 20% and they are found in all MPN subtypes. For comparison, TET2 mutations are present in 50% to 60% of chronic myelomonocytic leukemia (CMML).71 TET2 mutations can be also secondary mutations after JAK2V617F or CALR mutations and induce an expansion of double-mutated stem cells and progenitors.72 It has been suggested that when TET2 mutates first, the subsequent occurrence of JAK2V617F induces an ET phenotype, in contrast to the situation in which JAK2V617F mutates first and the phenotype is more likely PV. Secondary TET2 mutations can also be associated with progression, more particularly when the mutation is homozygous.73
DNMT3A mutations are less frequent than TET2 mutations in MPNs (5%-10%) and in the majority of the cases precede JAK2V617F or MPL mutations.74 As in other malignant hemopathies, the R882H is the most prevalent DNMT3A mutation. Both TET2 and DNMT3A mutations increase the self-renewal capacities of HSCs in both humans and mice.75,76 The precise mechanism is not completely understood, but TET2 and DNMT3A control the expression of genes involved in HSC properties as well as differentiation. It has been recently suggested that TET2-deficient HSCs are more prone to differentiation than normal HSCs, with the derepression of transcription factors such as Klf1.68
TET2 and DNMT3A are the 2 most frequently mutated genes associated with clonal hematopoiesis during aging.77-79 On the other hand, there is no clear evidence that these mutations induce gene instability. However, by inducing a clonal hematopoiesis and thus increasing replication of mutated stem cells, they may indirectly induce secondary mutations. They may also favor leukemogenesis by progressively modifying the equilibrium between self-renewal and differentiation. DNMT3A mutation may predispose to leukemic progression in MPNs. In mouse models, Tet2 loss increased the competitive reconstitution capacities of Jak2V617F-expressing HSCs and also worsened the MPN phenotype with an increase in extramedullary hematopoiesis without leukemic transformation.80,81
Overall, these 2 types of mutations increase the self-renewal of JAK2V617F HSCs and play an important role in disease initiation. However, they can also induce disease progression when they occur as secondary mutations, but their role in MF and leukemic transformation remains elusive.
Loss of EZH2 function promotes MF development and poor prognosis
EZH2 is 1 of the 2 histone methyltransferases of the PRC2 complex and is involved in the repressive H3K27 trimethylation (H3K27m3) (Figure 4). Loss-of-function mutations and cytogenetic lesions in EZH2 and other PRC2 members (SUZ12, JARID2, EED) have been described in MPNs and across all myeloid malignancies.82,83 EZH2 is mutated in 5% to 10% of PMF, more particularly associated with JAK2V617F and gives a poor prognosis.84 Three recent studies have shown that Ezh2 loss in a murine MPN model expressing JAK2V617F dramatically modifies the MPN phenotype. JAK2V617F/Ezh2−/− double mutant mice had significantly reduced survival due to rapid acceleration to lethal MF.85-87 Furthermore, strong cooperation between Ezh2 loss and JAK2V617F on disease initiation by increasing the fitness of JAK2V617F stem cells was reported. At the cellular level, Ezh2 loss increased megakaryopoiesis at the expense of erythropoiesis. The phenotype of the JAK2V617F/Ezh2−/− double mutant mice was related to changes in the expression of PRC2 target genes and more particularly of the Lin28b/let7/Hmga2 axis. The overexpression of Hmga2, a gene that was previously found deregulated in human PMF, appears to play an important role in this phenotype.86,87
ASXL1 and SRSF2 mutations are associated with a poor prognosis
ASXL1 encodes for an epigenetic regulator, which binds to chromatin. Interestingly, it has been shown that ASXL1 recruits the PRC2 complex to specific loci through a direct interaction between ASXL1 and EZH288 (Figure 4). However, the role of ASXL1 is wider than EZH2 because it can be also involved in the PRC1 complex by its association with the deubiquitinating enzyme (DUB) BRCA-1–associated protein (BAP1), a critical tumor suppressor in solid tumor.89 In addition, ASXL1 also binds to nuclear hormone receptors such as RARα and estrogen receptors. The ASXL family plays an important role in development as shown in Drosophila by regulating Hox genes. In humans, de novo or germ line ASXL1 mutations are responsible for a developmental syndrome called the Bohring-Opitz syndrome.90 Somatic mutations in ASXL1 were first described by Birnbaum’s group, first in MDS and CMML and then in MPN.69,91 They are the second mutations by their frequency in epigenetic regulators after TET2 in MPNs. However, they are essentially associated with PMF with a frequency around 25%. The ASXL1 mutations are associated with an aggravation in the prognosis, whatever the International Prognostic Scoring System (iPSS) classification. Mutations are loss of function and are associated with a higher frequency of AML transformation. These genetic alterations are either focal deletion or nonsense mutation or insertion/deletion leading to frameshift, the most common variant being the c.1934dupG;p.Gly646TrpfsX12 variant. In mice, Asxl1 deletion leads to profound cytopenia with a dysplasia associated with a profound defect in HSC self-renewal properties that are corrected by Tet2 deletion.92
SRSF2 mutations are also associated with MPNs having a poor prognosis.93 Mutations in genes encoding spliceosome proteins were detected in different hematopoietic malignancies in 2011, but more particularly in MDS.94 Spliceosome gene mutations in MPNs are essentially restricted to ET and MF. The most frequent mutations include SRSF2, SF3B1, and U2AF1 and are associated with anemia and thrombocytopenia.95 SRSF2 encodes for a protein, which is a member of the serine/arginine-rich splicing factor family that binds to exonic splicing enhancer (ESE) sequences in the pre–messenger RNA (mRNA). There is increasing evidence that SRSF2 mutations, essentially on the proline 95 are not loss of functions, but recognize preferentially the CCNG ESE motifs whereas the WT sequence recognizes both the CCNG and GGNG ESE motifs. This alters the splicing of numerous pre-mRNA and leads to numerous functionally relevant misspliced events. In consequence, mice with a heterozygous Srsf2P95H mutant develop a cytopenia by myelodysplasia whereas a homozygous Srsf2 knockout when induced in adults leads to cytopenia due to a decrease in hematopoietic progenitors associated with increased apoptosis mimicking a bone marrow aplasia.96 Particularly, it has been shown that the Srsf2 P95H mutation in mice can lead to a missplicing and nonsense-mediated decay of Ezh2.97 Knowing that SRSF2 mutations are enriched in leukemic transformation of MPNs (19% of the cases), this further establishes that EZH2 and the PRC2 complex are important factors in the development of MF and progression toward leukemia.
The p53 pathway and the development of secondary AML
Various mutations and cytogenetic lesions targeting the TP53 tumor-suppressor function are found in around 40% to 50% of secondary leukemia of MPNs. This usually includes missense mutations or deletions in the TP53 gene or amplification of chromosome 1q targeting MDM4, a TP53 transcriptional inhibitor.98 Heterozygous TP53-mutated clones can be found early in MPN, but evolution to leukemia is associated with its clonal dominance and transition from heterozygosity to homozygosity for the mutation.98 This mode of transformation mimics therapy-related leukemia and is quite different from sporadic leukemia.99 Presently, the only mouse model which leads to leukemia is the overexpression of JAK2V617F in a Tp53 knockout model.65 The role of therapy in the induction or on the selection of TP53-mutated clones is presently unknown. TP53 as well as DNMT3A mutations are apparently more frequent in post-PV or -ET AML than on post-MF leukemia.100 Other secondary leukemia are mainly associated with mutations in ASXL1, SRSF2, IDH1/2, CBL, and LNK. Late events include RUNX1 mutations (30% of the cases), FLT3–internal tandem duplication (15% of the cases), N-RAS, NF1, and deletion in IKZF1 and CUX1100,101 (Figure 4). The complex mutational profile of patients in the leukemic stage indicates that all patients at this stage are genetically unique with complex clonal hierarchies. Treatment is challenging and relapse almost always occurs.
Important factors for initiation
Aging
There is increasing evidence that JAK2V617F is relatively frequent in the aging healthy population and is presently estimated to be <0.5%.13 This indicates that JAK2V617F alone is sufficient to engender clonal hematopoiesis and suggests it has a disease-initiating role in MPN.
However, JAK2V617F being a frequent mutation, it has a low penetrance to give a MPN, suggesting that other factors must be associated to JAK2V617F to give rise to a disease. Aging is generally associated with a deregulation of HSCs, which lose their function and become myeloid-biased and less quiescent as a consequence of intrinsic and environmental changes. Overall, this suggests that JAK2V617F HSCs have higher competitive properties in this context, explaining why MPNs are age-related diseases.
Inflammation
All MPNs, particularly PMF, are associated with an important inflammatory response as demonstrated by the presence of high plasma levels of inflammatory cytokines as well as constitutional symptoms alleviated by anti-inflammatory therapies such corticoid and ruxolitinib.102 These inflammatory cytokines are synthetized by the mutated and nonmutated hematopoietic cells as well as by nonhematopoietic cells such as mesenchymal stromal cells.103 More particularly, it has been demonstrated that the JAK2V617F progenitors secrete IL1β, which induces the apopotic death of nestin+ cells providing a favorable environment for JAK2V617F HSCs.104 Moreover, it has been shown that tumor necrosis factor α (TNFα) gives a competitive advantage to JAK2V617F HSCs.105 IL33 may have similar properties.106 The inflammatory cytokines might also be involved in extramedullar hematopoiesis by favoring colonization of JAK2V617F HSCs and progenitors in the spleen.
Predisposition
Several susceptibility alleles that correspond to common polymorphisms in genes such as JAK2, particularly the JAK2 46/1 haplotype, TERT, MECOM, SH2B3, CHEK2, PINT, and GFI1B, may weakly (by twofold to sixfold) increase the development of the disease.13,107 The mechanism is presently unknown but these predisposition variants involve genes that play a role either in DNA damage response (CHEK2, TERT, JAK2 46/1 haplotype) or in the JAK2/STAT pathway. Recently, predisposition factors were discovered in the true MPN families with an autosomal pattern of inheritance but with an incomplete penetrance, suggesting that they are strong predisposing factors. Although RBBP6 mutations were found in MF families, a copy-number variation of 6 genes with 2 genes involved (ATG2B and GSKIP) was identified in MPN families with a particularly aggressive phenotype.108,109
Conclusions
The mutational landscape in classical MPN is much more complex than initially thought. However, abnormal activation of JAK2 is the common feature of disease, causing mutations in the 3 genes driving the MPNs. The genetic cause remains unknown in around 15% of chronic thrombocytosis classified as ET and in <10% of PMF. The rare triple-negative PMF appears more likely to be MDSs with secondary fibrosis. In the majority of ET and PV, only mutations in JAK2, CALR, and MPL can be found, suggesting that the deregulation in the cytokine receptor/JAK2 pathway is sufficient to induce a clonal myeloproliferative disorder reinforcing the concept that JAK2 is the main therapeutic target. The mechanism of JAK2 activation may define 2 main types of MPNs: (1) those driven by JAK2V61F, leading to ET, PV, and PMF and associated with proliferation and activation of the 3 main myeloid lineages (erythroid, granulocytic, and MK/platelet lineages) and (2) those driven by CALR and MPL mutants which are associated with ET and PMF and target essentially the MK/platelet lineage.
PMF represents a different disorder, which may be genetically closer to MDS, and its development may require the occurrence of several drivers preceding the JAK2V617F mutations and the presence of inflammation in the bone marrow microenvironment. The clinical heterogeneity seems to be mainly related to the combination of mutations and perhaps their order of acquisition. The precise role of each mutation and their impact on MPN phenotype will require further studies. Future work on the pathogenesis of MPNs will be more focused on the interaction between extrinsic factors and acquired genetic abnormalities in disease initiation and progression.
Acknowledgments
The authors are indebted to Isabelle Plo, Caroline Marty, and Stefan Constantinescu for their help to improve the manuscript.
This work was supported by grants from Ligue Nationale Contre le Cancer (“Equipe labellisée 2016”), from INSERM, and from Laboratory of Excellence Globule Rouge-Excellence. W.V. was funded by the program “Investissements d’avenir”. R.K. received funding from the Austrian Science Fund (SFB F4702 and P29018-B30).
Authorship
Contribution: W.V. and R.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William Vainchenker, INSERM UMR 1170, Institut Gustave Roussy, 39, Rue Camille Desmoulins, Villejuif, 94805 France; e-mail: verpre@igr.fr; and Robert Kralovics, Research Center for Molecular Medicine of the Austrian Academy of Sciences, Lazarettgasse 14, AKH BT 25.3, A-1090 Vienna, Austria; e-mail: rkralovics@cemm.at.


![Figure 3. MKs play a central role in MPN pathogenesis. The MPL/JAK2 pathway is activated by the 3 MPN-restricted mutations (JAK2V617F, CALR mutants, and MPL mutants) placing MK hyperplasia and eventually dysplasia as a central determinant in MPN. MKs are mainly involved in platelet production, and also play an important role in the hematopoietic niche by regulating HSCs, remodeling the marrow by secretion of TGF-β1 and other cytokines (platelet-derived growth factor [PDGF], vascular endothelial growth factor [VEGF]) which ultimately lead to MF, and also inducing a neo-osteogenesis by inducing an osteoblastic differentiation through TGF-β1 and inhibiting osteoclast differentiation through osteoprotegerin. Furthermore, MKs secrete numerous inflammatory cytokines such as IL1α. The mechanisms of local activation of TGF-β1 are poorly known. OPG, osteoprotegerin; RANK, receptor activator of NF-κB; RANKL, RANK ligand; TGF, transforming growth factor; TSP, thrombospondin. Professional illustration by Somersault18:24.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/6/10.1182_blood-2016-10-695940/4/m_blood695940f3.jpeg?Expires=1767710213&Signature=T3MTybkA8PwSFFLYZubxhEqQ25~vGaiVE514vq8FCq~ubv9d6LsvLTE3w2GBUn4sYOswPcocrYcNQc2h1t8Dgka6xVTg5n0Zkv~QHLUKBpU8ffe2f~fzLQyFai1iavNlVRhtMQ0u0vnrhO~bpKGXBBYUUUZblcMPneVw70RzJZOlJPSGxb8WYFXeiG-L6VlhNd-PNPAK3JS-Ou6Yb1~XFu6G5cZ2zqN8nk5VyQDeVt0uOd7ql8m-9vHtnnYwbxLQpW7bO5b4jVT5VM1AiID2IMWEVzFef6ROHuiIgcm99~SWm-0EWXX~IDpv3QNSA5u0dkb4tfg167g5EN2JMC0tLw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

