Key Points
Our study identified a region on chromosome 6 comprising the genes SMAP1, B3GAT2, and RIMS1 as novel susceptibility locus for pediatric VTE.
Nonsynonymous variants in SMAP1 and RIMS1 are predicted as deleterious and may influence vesicle processing in blood cells.
Abstract
Recent genome-wide association studies (GWAS) have confirmed known risk mutations for venous thromboembolism (VTE) and identified a number of novel susceptibility loci in adults. Here we present a GWAS in 212 nuclear families with pediatric VTE followed by targeted next-generation sequencing (NGS) to identify causative mutations contributing to the association. Three single nucleotide polymorphisms (SNPs) exceeded the threshold for genome-wide significance as determined by permutation testing using 100 000 bootstrap permutations (P < 10−5). These SNPs reside in a region on chromosome 6q13 comprising the genes small ARF GAP1 (SMAP1), an ARF6 guanosine triphosphatase-activating protein that functions in clathrin-dependent endocytosis, and β-1,3-glucoronyltransferase 2 (B3GAT2), a member of the human natural killer 1 carbohydrate pathway. Rs1304029 and rs2748331 are associated with pediatric VTE with unpermuted/permuted values of P = 1.42 × 10−6/2.0 × 10−6 and P = 6.11 × 10−6/1.8 × 10−5, respectively. Rs2748331 was replicated (P = .00719) in an independent study sample coming from our GWAS on pediatric thromboembolic stroke (combined P = 7.88 × 10−7). Subsequent targeted NGS in 24 discordant sibling pairs identified 17 nonsynonymous coding variants, of which 1 located in SMAP1 and 3 in RIMS1, a member of the RIM family of active zone proteins, are predicted as damaging by Protein Variation Effect Analyzer and/or sorting intolerant from tolerant scores. Three SNPs curtly missed statistical significance in the transmission-disequilibrium test in the full cohort (rs112439957: P = .08326, SMAP1; rs767118962: P = .08326, RIMS1; and rs41265501: P = .05778, RIMS1). In conjunction, our data provide compelling evidence for SMAP1, B3GAT2, and RIMS1 as novel susceptibility loci for pediatric VTE and warrant future functional studies to unravel the underlying molecular mechanisms leading to VTE.
Introduction
Venous thromboembolism (VTE) is a common multifactorial disease that is influenced by environmental and genetic factors. Meta-analyses suggest that inherited thrombophilic mutations in coagulation factors identified in adult patients (ie, Factor VLeiden [FV]; prothrombin, deficiency of protein S, protein C, and antithrombin; and fibrinogen α [FGA] and fibrinogen γ [FGG] chains) are also the most common genetic determinants of pediatric first VTE and ischemic stroke.1-6 In recent years, novel loci have been suggested to confer susceptibility to VTE in adults through genome-wide association studies (GWAS). In 2009, Trégouët and colleagues reported the first GWAS, which confirmed known genetic susceptibility factors, FV and the ABO locus, for VTE in adults, but failed to identify novel susceptibility genes at genome-wide significance.7 In a follow-up study, using a collection of 5862 cases for VTE and 7112 healthy controls, Morange and colleagues8 implicated the HIVEP1 locus as a novel VTE susceptibility locus, which is not involved in the traditional coagulation system. A study by Heit and colleagues confirmed the significant association with FV and ABO, and implicated 2 potential novel susceptibility genes (BLZF1, NME7) for VTE in adults.9 Recently, a meta-analysis comprising 12 GWAS totaling 7507 VTE case subjects and 52 632 control subjects revealed 9 associated loci with genome-wide significance, confirming 6 known loci (F2, F5, F11, FGG, ABO, PROCR) and adding 3 new loci to the list of potential candidates (ZFPM2, TSPAN15, SLC44A2).10 However, a large proportion of missing heritability remains to be accounted for.10,11
To date, studies in families with a known first onset of pediatric VTE are lacking mainly because of limited sample size. VTE in children is a rare event and most likely to occur in neonates and children with underlying conditions such as cancer, sepsis, autoimmune disease, and congenital heart disease, or as a consequence of invasive therapeutic procedures. Prominent complicating features of VTE in children include lack of thrombus resolution and development of postthrombotic syndrome.1,2,4 Because of the low incidence of pediatric VTE in the general population (0.07-0.14/10 000 children; ∼5/10 000 hospital admissions of children), studies addressing the impact of genetic modifiers of thrombosis on the incidence of pediatric VTE are limited by the relatively small number of patients and often remain inconclusive. Therefore, we performed a family-based GWAS for pediatric VTE in 212 trios followed by a targeted next-generation sequencing (NGS) approach in 48 children to identify rare coding variants in the genes underlying the association signal.
Methods
Subjects
Plasma and DNA samples of 322 families of children with VTE (stage I) (ie, samples of the propositus, nonaffected brothers or sisters and biological parents) were collected between January 2002 and December 2007. All probands were of Caucasian descent. The present study was performed in accordance with the ethical standards in the updated version of the 1964 Declaration of Helsinki and was approved by the medical ethics committee of the University of Muenster, Germany. With written parental consent, term neonates and children with confirmed diagnosis of VTE not older than 18 years at onset, biological brothers and sisters, and available parents were enrolled. Premature birth (≤36 gestational weeks), patients older than 18 years at onset, children with a first ischemic stroke of vascular origin, children with missing parents, and families with nonpaternity were excluded. Further exclusion criteria were ongoing liver, renal, or inflammatory diseases; malignancies; and concurrent treatment regimens known to influence fibrinogen levels. From the initial study sample, 212 affected offspring trios were subject to genome-wide genotyping and statistical analysis.
The stage II replication study sample comprises 201 Caucasian families of children with thromboembolic stroke (TS) of cardiac origin or resulting from paradoxical embolism that have been described in a previous study. Selection criteria and sample characteristics for affected offspring trios are given there in detail.12 In brief, stroke subtypes of the children enrolled were reclassified according to explicit predefined Trial of ORG 10172 in Acute Stroke Treatment criteria modified for children. Pediatric acute ischemic stroke (AIS) not classified as arteriopathy (ie, AIS of cardiac origin or AIS resulting from paradoxical embolism) was considered to be thromboembolic AIS. Children with arteriopathy, moyamoya arteriopathy, Ehlers-Danlos syndrome, Marfan syndrome, or α1-antitrypsin deficiency were not enrolled. Preterm AIS infants, perinatal stroke patients, and patients >18 years of age at first AIS onset were also not included. In addition, patients with suspected AIS without the diagnosis being confirmed by an independent experienced pediatric neuroradiologist were excluded from the survey.
The stage III replication study cohort consisted of 651 consecutively enrolled adult patients of Caucasian descent with objectively confirmed VTE ascertained between December 2010 and July 2013 from Hamburg, Schleswig-Holstein, and Mecklenburg-Vorpommern in the northern part of Germany. Patients with cancer or pregnancy-related VTE were excluded from ascertainment. In addition, 1356 individuals enrolled in the population-based recruitment for genetics research (PopGen) population13 were used as controls.
An overview of sample characteristics for all stages is given in supplemental Tables 1 and 2, available on the Blood Web site.
Genotyping
In the initial analysis, 212 trios were genotyped on the Illumina Human660W-Quad v1.0 BeadChips (Illumina, Eindhoven, NL). Potential population stratification was assessed using a population concordance analysis implemented in PLINK version 1.07.14 The stage II replication sample comprising 201 trios with pediatric TS were previously genotyped on the Illumina Infinium 370 CNV SNP.12 The stage III replication sample comprising 651 adult VTE cases and 1356 PopGen controls were genotyped for rs1304029 and rs2748331 using the TaqMan allelic discrimination method (assay from Life Technologies, Darmstadt, Germany) on an ABI 7900HT sequence detector (Applied Biosystems, Darmstadt, Germany) using 2 ng of genomic DNA per SNP. Genotypes were generated by automatic calling using SDS software, version 2.3 (Applied Biosystems).
Imputation of SNPs in target region
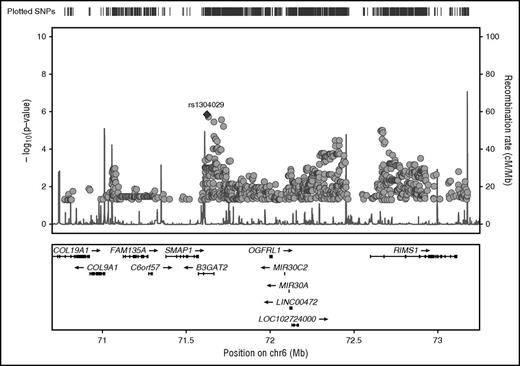
Before imputation of SNPs in our target region on chromosome 6, we excluded SNPs based on a missing rate >2% and aligned all genotypes to the forward strand. The remaining 543 GWAS SNPs were used for imputation. A 2-step procedure was used to impute unobserved genotypes using phased haplotypes from the integrated phase 3 release of the 1000 Genomes Project. In a first prephasing step, we used SHAPEIT15 to estimate haplotypes for the VTE study samples. In a second step, we imputed missing alleles for additional SNPs directly onto these phased haplotypes using IMPUTE2.16 Imputed SNPs with high confidence (maximum probability ≥0.90) were selected for plotting in Figure 2.
Association analysis
The following quality control thresholds were used for the genome-wide analysis of the stage I VTE study sample: calling fraction per chip ≥.90, minor allele frequency ≥0.01, P (deviation from Hardy-Weinberg equilibrium) ≥ .001, missing frequency per SNP <0.05, Mendel error (family-wise) <0.05, and Mendel error (SNP-wise) <0.1. Estimated gender matches as computed by GenomeStudio Software (Illumina Inc. V1.9.4).
Subsequent to quality control, the dataset consists of 212 trios and 529 944 SNPs and exhibits a call rate >99%. We performed a GWAS using a threshold of P <1 × 10−5 for suggestive genome-wide association. P values for single-point and haplotype associations were computed using the transmission-disequilibrium test17 and validated by up to 1 000 000 gene-dropping permutation runs in an adaptive approach yielding empirical P values based on unbiased null distributions as implemented in PLINK, version 1.07.14 No adjustment for covariates was applied because of the family design of the study.
SNPs exceeding the threshold of P <1 × 10−5 in stage I were assessed concerning their validity by independent replication in 201 trios with pediatric TS (stage II).12 A nominal P < .05 was considered significant for replication purposes. A combined P value for stage I (VTE) and stage II (TS) association was calculated using the method by Fisher.18
The 2 most significant SNPs—rs1304029 and rs2748331—implicated from the pediatric study sample were replicated in stage III in 651 adult cases and 1356 PopGen controls using a case-control design. Association was tested in PLINK using logistic regression for the 3 modes of inheritance (recessive, dominant, additive) and adjusted for gender and multiple testing using the Bonferroni correction.
Targeted resequencing
A subset of 24 independent pediatric probands from the stage I study sample and 24 unaffected siblings were processed for targeted resequencing. Probands were selected for sequencing if an unaffected sibling had been enrolled in our study and sufficient DNA material was available for both. Genomic DNA samples of all samples were quantified using the Qubit dsDNA assay kit (Thermo Fisher Scientific, Waltham, MA). Following the SureSelectQXT protocol for custom target enrichment (Agilent Technologies, Santa Clara, CA), library construction was carried out using 50 ng of genomic DNA. The captured genomic region (2.42 Mb) encompasses the genomic region from 70 911 768 to 73 220 274 bp on chromosome 6 (hg19). The 48 sequencing libraries were quality controlled and quantified using the Bioanalyzer High Sensitivity DNA Analysis Kit (Agilent Technologies) and the KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, DE). The equimolar library pool was clustered and sequenced using the Illumina MiSeq Reagent Kit v3 on an Illumina MiSeq sequencing system in 3 dual index-paired end-sequencing runs (75 cycles).
Initial quality filtering, read mapping, and variant calling
Sequencing reads were subject to quality control (trimming adapter sequences from the raw reads, discarding low-quality bases at the reads' ends) using an in-house–developed software and were mapped to the reference genome (hg19) using the BWA software tool (version 0.6.2).19 More than 99% of the reads were mapped successfully. The mean coverage of the target region was approximately 40.
The Picard software tool (version 1.73, http://broadinstitute.github.io/picard) was used to remove duplicated reads from further analysis. Reads were realigned around indels and quality score recalibration was performed using GATK (version 3.4).20-22 After variant calling using GATK tool UnifiedGenotyper, both SNP and indel calls were filtered for low coverage depth (depth of coverage <5). Individual genotypes with genotype quality values reported by the GATK engine below a threshold of 10 for SNPs and 20 for indels were considered as unknown (no-call). After applying these filtering criteria, we identified an 8742 SNPs and 1619 indels in the target region. To assess a potential functional effect of genetic variants, we used the SnpEff software tool (version 4.1)23 and Annovar (version 2015Jun23).24
SDT and rare variant analysis for family-based design
We used a custom software implementation of the sibship disequilibrium test (SDT)25 for association analysis of detected variants in the sample of 24 discordant sibling pairs.
De et al (2013) propose a method to test for association of rare variants obtained by sequencing in family-based samples by collapsing the standard family-based association test (FBAT) statistic. They also propose a suitable weighting scheme so that low-frequency SNPs that may be enriched in functional variants can be up-weighted compared with common variants.26 We used this weighted rare variant statistics as implemented in the FBAT software, tool 2.0.4.27 SNPs were selected if they are present in at least 1 of the individuals and having minor allele frequency <3%.
Validation of variants through capillary sequencing and genotyping in full cohort
Variant validation was performed through direct DNA sequencing using Big-Dye Terminator, version 3.1, Cycle Sequencing Kit (Life Technologies) on a 3730 DNA Analyzer (Life Technologies). Variant regions were amplified using AmpliTaq Gold PCR Master Mix (Life Technologies) or Phusion High-Fidelity PCR Master Mix (NEB, Frankfurt, Germany), respectively. ExoSap-IT kit (Affymetrix, Santa Clara, CA) was used for PCR product cleanup. Sequence analysis was performed using CodonCode Aligner (CodonCode Corporation, Centerville, MA). Validated variants were subsequently genotyped in the stage I study sample using KASP Genotyping Assays (LGC Limited, Leeds, United Kingdom) on an ABI 7900HT sequence detector (Applied Biosystems) using 2 ng of genomic DNA per SNP. Genotype calling and association analysis was performed as described previously.
Results
GWAS and replication study
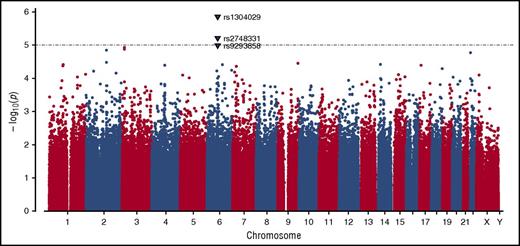
GWAS was conducted in the complete dataset of 212 incident VTE families genotyped using the Infinium 660W SNP array. The average genotype call rate was >99% and the genomic inflation factor was λ = 1.017, indicating the absence of population stratification. A total of 529 944 SNPs provided valid association signals after applying the quality control criteria indicated previously, resulting in a Manhattan plot as shown in Figure 1. Two SNPs exceeded the threshold for genome-wide significance in this dataset as determined by permutation testing using up to 1 000 000 bootstrap permutations (P < 10−5), and are likely true associations (Table 1). One of these SNPs (rs1304029) and another curtly missing the mentioned threshold for permuted P value (rs2748331) reside in a region on chromosome 6q13 that comprises 2 overlapping genes: small ARF GAP1 (SMAP1), an ARF6 guanosine triphosphatase (GTPase)-activating protein that functions in clathrin-dependent endocytosis, and β-1,3-glucoronyltransferase 2 (B3GAT2), a member of the human natural killer-1 (HNK-1) carbohydrate pathway (Figure 2). These SNPs are associated with pediatric VTE with unpermuted/permuted values for rs1304029 (P = 1.42 × 10−6/P = 2.00 × 10−6) and rs2748331 (P = 6.11 × 10−6/P = 1.8 × 10−5). The corresponding haplotype association test resulted in a P = 2.76 × 10−6 for the AG haplotype, further underlining the robustness of the association.
Manhattan plot demonstrating the distribution of P values of stage I association analysis. The y-axis shows the unpermuted −log10P values of 529 944 SNPs and the x-axis depicts their chromosomal positions. The P value threshold of 10−5 is indicated by a gray dashed horizontal line. Variants associated with P values below this threshold are highlighted as triangles and annotated with their corresponding SNP identification numbers.
Manhattan plot demonstrating the distribution of P values of stage I association analysis. The y-axis shows the unpermuted −log10P values of 529 944 SNPs and the x-axis depicts their chromosomal positions. The P value threshold of 10−5 is indicated by a gray dashed horizontal line. Variants associated with P values below this threshold are highlighted as triangles and annotated with their corresponding SNP identification numbers.
SNPs associated at permuted P < 1 × 10−4 with pediatric VTE in the GWAS (stage I) after up to 1 000 000 bootstrap mutations
| Variant . | Chr . | Gene . | Alt . | Ref . | MAF . | T . | U . | OR (95% CI) . | P . | P (perm) . | No. of perm . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1304029 | 6 | B3GAT2 | A | G | 0.3667 | 63 | 130 | 0.4846 (0.3587-0.6547) | 1.42e-06 | 2.00e-06 | 1 000 000 |
| rs9293858 | 6 | RIMS1 | C | T | 0.2323 | 50 | 105 | 0.4762 (0.34-0.6669) | 9.98e-06 | 8.00e-06 | 1 000 000 |
| rs2748331 | 6 | B3GAT2 | G | A | 0.3042 | 58 | 118 | 0.4915 (0.359-0.673) | 6.11e-06 | 1.80e-05 | 1 000 000 |
| rs914958 | 1 | ABCA4 | G | A | 0.2323 | 51 | 102 | 0.5 (0.3573-0.6998) | 3.74e-05 | 1.80e-05 | 1 000 000 |
| rs4529013 | 4 | MAPK10 | T | C | 0.3267 | 62 | 117 | 0.5299 (0.3895-0.721) | 3.94e-05 | 2.00e-05 | 1 000 000 |
| rs9957519 | 18 | LOC105372224 | T | G | 0.1686 | 39 | 84 | 0.4643 (0.3176-0.6788) | 4.96e-05 | 2.10e-05 | 1 000 000 |
| rs1865590 | 2 | THSD7B | C | T | 0.3002 | 118 | 60 | 1.967 (1.441-2.683) | 1.38e-05 | 2.40e-05 | 1 000 000 |
| rs9606534 | 22 | IGKV2OR22-4 | G | A | 0.1509 | 34 | 80 | 0.425 (0.2845-0.6348) | 1.65e-05 | 3.30e-05 | 1 000 000 |
| rs11128790 | 3 | RFTN1 | A | G | 0.112 | 59 | 20 | 2.95 (1.777-4.899) | 1.15e-05 | 3.40e-05 | 1 000 000 |
| rs4792119 | 17 | SHISA6 | C | T | 0.2193 | 54 | 106 | 0.5094 (0.3671-0.707) | 3.94e-05 | 3.50e-05 | 1 000 000 |
| rs9399770 | 6 | LOC100996860 | T | G | 0.3667 | 73 | 132 | 0.553 (0.4155-0.736) | 3.78e-05 | 4.00e-05 | 1 000 000 |
| rs17576372 | 1 | TGFBR3 | G | A | 0.3962 | 125 | 68 | 1.838 (1.368-2.47) | 4.08e-05 | 4.57e-05 | 897 000 |
| rs10247053 | 7 | LOC105375203 | A | G | 0.2854 | 60 | 114 | 0.5263 (0.385-0.7195) | 4.25e-05 | 5.35e-05 | 767 000 |
| rs636434 | 6 | EYS | A | G | 0.3432 | 129 | 72 | 1.792 (1.343-2.39) | 5.81e-05 | 5.35e-05 | 766 000 |
| rs657152 | 9 | ABO | A | C | 0.4269 | 143 | 81 | 1.765 (1.344-2.319) | 3.44e-05 | 5.98e-05 | 686 000 |
| rs10190178 | 2 | THSD7B | A | G | 0.2998 | 115 | 60 | 1.917 (1.403-2.619) | 3.22e-05 | 6.15e-05 | 667 000 |
| rs5014872 | 2 | LOC730100 | C | A | 0.1584 | 38 | 82 | 0.4634 (0.3154-0.6808) | 5.90e-05 | 6.21e-05 | 660 000 |
| rs3823606 | 7 | TPST1 | A | G | 0.0196 | 0 | 15 | NA | 1.08e-04 | 6.27e-05 | 654 345 |
| rs10498910 | 6 | LOC105377862 | G | A | 0.1427 | 75 | 34 | 2.206 (1.471-3.308) | 8.60e-05 | 6.89e-05 | 595 000 |
| rs1565242 | 15 | LOC105370983 | G | A | 0.1368 | 32 | 72 | 0.4444 (0.2931-0.674) | 8.77e-05 | 7.23e-05 | 567 000 |
| rs1958059 | 14 | NPAS3 | T | C | 0.1686 | 37 | 82 | 0.4512 (0.3061-0.6652) | 3.71e-05 | 7.28e-05 | 563 000 |
| rs1521882 | 2 | KIAA2012 | C | T | 0.1934 | 83 | 39 | 2.128 (1.455-3.114) | 6.79e-05 | 7.48e-05 | 548 000 |
| rs17781793 | 12 | MRPL40P1 | G | A | 0.1223 | 20 | 53 | 0.3774 (0.2256-0.6311) | 1.12e-04 | 7.81e-05 | 525 000 |
| rs4775384 | 15 | RORA | T | C | 0.1274 | 25 | 61 | 0.4098 (0.2573-0.6528) | 1.04e-04 | 8.16e-05 | 502 497 |
| rs1948650 | 15 | DPH6-AS1 | T | C | 0.3297 | 112 | 61 | 1.836 (1.344-2.508) | 1.06e-04 | 8.71e-05 | 471 000 |
| rs436985 | 5 | C5orf64 | G | A | 0.395 | 78 | 135 | 0.5778 (0.4372-0.7635) | 9.40e-05 | 9.13e-05 | 449 000 |
| rs4926448 | 1 | SCCPDH | C | T | 0.4575 | 78 | 136 | 0.5735 (0.4342-0.7576) | 7.35e-05 | 9.38e-05 | 437 124 |
| rs11153626 | 6 | FAM162B | T | C | 0.2748 | 107 | 58 | 1.845 (1.34-2.54) | 1.36e-04 | 9.49e-05 | 432 000 |
| rs2214810 | 7 | LOC101928077 | G | T | 0.2854 | 61 | 113 | 0.5398 (0.3954-0.737) | 8.08e-05 | 9.62e-05 | 436 563 |
| Variant . | Chr . | Gene . | Alt . | Ref . | MAF . | T . | U . | OR (95% CI) . | P . | P (perm) . | No. of perm . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1304029 | 6 | B3GAT2 | A | G | 0.3667 | 63 | 130 | 0.4846 (0.3587-0.6547) | 1.42e-06 | 2.00e-06 | 1 000 000 |
| rs9293858 | 6 | RIMS1 | C | T | 0.2323 | 50 | 105 | 0.4762 (0.34-0.6669) | 9.98e-06 | 8.00e-06 | 1 000 000 |
| rs2748331 | 6 | B3GAT2 | G | A | 0.3042 | 58 | 118 | 0.4915 (0.359-0.673) | 6.11e-06 | 1.80e-05 | 1 000 000 |
| rs914958 | 1 | ABCA4 | G | A | 0.2323 | 51 | 102 | 0.5 (0.3573-0.6998) | 3.74e-05 | 1.80e-05 | 1 000 000 |
| rs4529013 | 4 | MAPK10 | T | C | 0.3267 | 62 | 117 | 0.5299 (0.3895-0.721) | 3.94e-05 | 2.00e-05 | 1 000 000 |
| rs9957519 | 18 | LOC105372224 | T | G | 0.1686 | 39 | 84 | 0.4643 (0.3176-0.6788) | 4.96e-05 | 2.10e-05 | 1 000 000 |
| rs1865590 | 2 | THSD7B | C | T | 0.3002 | 118 | 60 | 1.967 (1.441-2.683) | 1.38e-05 | 2.40e-05 | 1 000 000 |
| rs9606534 | 22 | IGKV2OR22-4 | G | A | 0.1509 | 34 | 80 | 0.425 (0.2845-0.6348) | 1.65e-05 | 3.30e-05 | 1 000 000 |
| rs11128790 | 3 | RFTN1 | A | G | 0.112 | 59 | 20 | 2.95 (1.777-4.899) | 1.15e-05 | 3.40e-05 | 1 000 000 |
| rs4792119 | 17 | SHISA6 | C | T | 0.2193 | 54 | 106 | 0.5094 (0.3671-0.707) | 3.94e-05 | 3.50e-05 | 1 000 000 |
| rs9399770 | 6 | LOC100996860 | T | G | 0.3667 | 73 | 132 | 0.553 (0.4155-0.736) | 3.78e-05 | 4.00e-05 | 1 000 000 |
| rs17576372 | 1 | TGFBR3 | G | A | 0.3962 | 125 | 68 | 1.838 (1.368-2.47) | 4.08e-05 | 4.57e-05 | 897 000 |
| rs10247053 | 7 | LOC105375203 | A | G | 0.2854 | 60 | 114 | 0.5263 (0.385-0.7195) | 4.25e-05 | 5.35e-05 | 767 000 |
| rs636434 | 6 | EYS | A | G | 0.3432 | 129 | 72 | 1.792 (1.343-2.39) | 5.81e-05 | 5.35e-05 | 766 000 |
| rs657152 | 9 | ABO | A | C | 0.4269 | 143 | 81 | 1.765 (1.344-2.319) | 3.44e-05 | 5.98e-05 | 686 000 |
| rs10190178 | 2 | THSD7B | A | G | 0.2998 | 115 | 60 | 1.917 (1.403-2.619) | 3.22e-05 | 6.15e-05 | 667 000 |
| rs5014872 | 2 | LOC730100 | C | A | 0.1584 | 38 | 82 | 0.4634 (0.3154-0.6808) | 5.90e-05 | 6.21e-05 | 660 000 |
| rs3823606 | 7 | TPST1 | A | G | 0.0196 | 0 | 15 | NA | 1.08e-04 | 6.27e-05 | 654 345 |
| rs10498910 | 6 | LOC105377862 | G | A | 0.1427 | 75 | 34 | 2.206 (1.471-3.308) | 8.60e-05 | 6.89e-05 | 595 000 |
| rs1565242 | 15 | LOC105370983 | G | A | 0.1368 | 32 | 72 | 0.4444 (0.2931-0.674) | 8.77e-05 | 7.23e-05 | 567 000 |
| rs1958059 | 14 | NPAS3 | T | C | 0.1686 | 37 | 82 | 0.4512 (0.3061-0.6652) | 3.71e-05 | 7.28e-05 | 563 000 |
| rs1521882 | 2 | KIAA2012 | C | T | 0.1934 | 83 | 39 | 2.128 (1.455-3.114) | 6.79e-05 | 7.48e-05 | 548 000 |
| rs17781793 | 12 | MRPL40P1 | G | A | 0.1223 | 20 | 53 | 0.3774 (0.2256-0.6311) | 1.12e-04 | 7.81e-05 | 525 000 |
| rs4775384 | 15 | RORA | T | C | 0.1274 | 25 | 61 | 0.4098 (0.2573-0.6528) | 1.04e-04 | 8.16e-05 | 502 497 |
| rs1948650 | 15 | DPH6-AS1 | T | C | 0.3297 | 112 | 61 | 1.836 (1.344-2.508) | 1.06e-04 | 8.71e-05 | 471 000 |
| rs436985 | 5 | C5orf64 | G | A | 0.395 | 78 | 135 | 0.5778 (0.4372-0.7635) | 9.40e-05 | 9.13e-05 | 449 000 |
| rs4926448 | 1 | SCCPDH | C | T | 0.4575 | 78 | 136 | 0.5735 (0.4342-0.7576) | 7.35e-05 | 9.38e-05 | 437 124 |
| rs11153626 | 6 | FAM162B | T | C | 0.2748 | 107 | 58 | 1.845 (1.34-2.54) | 1.36e-04 | 9.49e-05 | 432 000 |
| rs2214810 | 7 | LOC101928077 | G | T | 0.2854 | 61 | 113 | 0.5398 (0.3954-0.737) | 8.08e-05 | 9.62e-05 | 436 563 |
Alt, alternative allele; Chr, chromosome; NA, not available; no. of perm, number of applied permutations; P (perm.), empirical P value by permutations; Ref, reference allele; T, number of transmitted minor alleles; U, number of untransmitted minor alleles.
Regional plot of chromosome 6 target region with association P values of stage I analysis. GWAS SNPs and imputed SNPs with association P values <.05 are plotted in their genomic context using LocusZoom. SNP rs1304029 is highlighted by a filled diamond symbol.
Regional plot of chromosome 6 target region with association P values of stage I analysis. GWAS SNPs and imputed SNPs with association P values <.05 are plotted in their genomic context using LocusZoom. SNP rs1304029 is highlighted by a filled diamond symbol.
The association with B3GAT2 was replicated for SNPs rs2748331 in an independent study sample (stage II) of 201 families for pediatric TS (rs2748331: P = .00719), resulting in a combined value of P = 7.88 × 10−7. Another B3GAT2 SNP, rs9446340, which was associated with an unpermuted value of P = .015 in stage I, was replicated in stage II with P = .01, resulting in a combined value of P = 1.48 × 10−3. Twenty-seven additional SNPs were associated at confident permuted values (P <1 × 10−4) in stage I, but failed to exceed the threshold for genome-wide significance (P <1 × 10−5) (Table 1). From these, rs10498910 (stage I values unpermuted/permuted: P = 8.60 × 10−5/P = 6.89 × 10−5) mapped to a hypothetical gene (LOC105377862) on chromosome 6 about 6 Mb apart from B3GAT2, was replicated in our stage II sample (rs10498910: P = .05; combined P = 5.742 × 10−5).
In stage III, in a sample including 651 adults affected with VTE and 1356 healthy controls, the 2 SNPs, rs1304029 and rs2748331, located within or next to SMAP1/B3GAT2 coming from the pediatric VTE GWAS, were significantly associated with adult VTE in the additive and dominant genotype models: rs1304029, P = .027; odds ratio (OR), 1.18 (95% confidence interval [CI], 1.02-1.36) (additive) and P = .017; OR, 1.28 (95% CI, 1.04-1.56) (dominant); rs2748331: P = .024; OR, 1.20 (95% CI, 1.02-1.40) (additive), P = .009; OR, 1.30 (95% CI, 1.07-1.58) (dominant) at nominal P values <.05. However, after Bonferroni correction for multiple testing, these P values became nominally insignificant and thus need to be viewed with caution.
Overlap with VTE susceptibility genes from GWAS in adults
Among the previously reported loci,1-7 the ABO locus provided the strongest association signals for 3 SNPs—rs495828 (P = 6.44 × 10−4), rs505922 (P = 4.03 × 10−4), and rs657152 (P = 3.44 × 10−5)—almost reaching genome-wide significance. In addition, rs13146272 located in CYP4V2 (MIM 608614) (P = 9.58 × 10−4) and rs925451 in F11 (P = 2.76 × 10−3) were significantly associated with VTE in children. None of the other loci implicated by previous GWAS in adults was replicated in our study sample for pediatric VTE.
Targeted NGS and validation
The captured region on chromosome 6 (2.42 Mbp) contains the following genes: COL19A1, COL9A1, EVADR, FAM135A, SDHAF4, SMAP1, B3GAT2, OGFRL1, MIR30C2, MIR30A, LINC00472, LINC01626, and RIMS1. The quality of the alignment sequencing data showed a mapping rate >99%, a mean mapping quality of 45.4, and a mean coverage of 39.9. After variant calling and quality filtering, we identified 8742 SNPs and 1619 indels overall in the target region on chromosome 6, of which 850 SNPs and 741 indels are novel. Seventeen nonsynonymous coding variants (14 SNPs and 3 indels) in the genes C6orf57, COL9A1, FAM135A, RIMS1, and SMAP1 are shown in Table 2. The variant count per sample for nonsynonymous SNPs ranged from 1 to 7. Seven nonsynonymous variants showing either a Protein Variation Effect Analyzer score below −2.5 and/or sorting intolerant from tolerant score <0.05 have been predicted as damaging (supplemental Table 3). Thirteen SNPs chosen for validation by capillary sequencing showed identical results to the NGS calls.
Exonic SNPs and indels of target region on chromosome 6 detected by NGS in 24 discordant VTE sibling pairs
| Variant . | BP . | Gene . | Alt . | Ref . | MAF . | Effect . | Geno . | AA change . |
|---|---|---|---|---|---|---|---|---|
| rs6910140 | 70944257 | COL9A1 | C | T | 0.013 | Nonsyn | 47/1/0 | COL9A1:NM_078485:exon29:c.A1570G:p.M524V,COL9A1:NM_001851:exon35:c.A2299G:p.M767V |
| rs1135056 | 70961833 | COL9A1 | C | T | 0.407 | Nonsyn | 20/22/6 | COL9A1:NM_078485:exon22:c.A1133G:p.Q378R,COL9A1:NM_001851:exon28:c.A1862G:p.Q621R |
| rs592121 | 70984436 | COL9A1 | G | A | 0.367 | Nonsyn | 22/19/7 | COL9A1:NM_078485:exon5:c.T286C:p.S96P,COL9A1:NM_001851:exon11:c.T1015C:p.S339P |
| rs2747701 | 71238105 | FAM135A | G | A | 0.451 | Nonsyn | 19/17/12 | FAM135A:NM_001162529:exon14:c.A3725G:p.D1242G,FAM135A:NM_020819:exon15:c.A3086G:p.D1029G,FAM135A:NM_001105531:exon16:c.A3137G:p.D1046G |
| rs1048886 | 71289189 | C6orf57 | G | A | 0.206 | Nonsyn | 31/14/3 | C6orf57:NM_145267:exon2:c.A137G:p.Q46R |
| rs146446063 | 71298323 | C6orf57 | T | C | 0.007 | Nonsyn | 44/4/0 | C6orf57:NM_145267:exon3:c.C223T:p.P75S |
| rs112439957 | 71377781 | SMAP1 | A | C | 0.008 | Nonsyn | 44/4/0 | SMAP1:NM_001044305:exon1:c.C55A:p.L19I,SMAP1:NM_001281439:exon1:c.C55A:p.L19I,SMAP1:NM_021940:exon1:c.C55A:p.L19I |
| rs200042613 | 71378354 | SMAP1 | A | AC | 0.033 | Frameshift deletion | 45/3/0 | SMAP1:NM_001281440:exon1:c.24delC:p.H8fs |
| rs2273566 | 71546702 | SMAP1 | T | C | 0.084 | Nonsyn | 40/8/0 | SMAP1:NM_001281439:exon6:c.C554T:p.A185V,SMAP1:NM_021940:exon6:c.C554T:p.A185V,SMAP1:NM_001044305:exon7:c.C635T:p.A212V,SMAP1:NM_001281440:exon7:c.C605T:p.A202V |
| rs201201431 | 71569066 | SMAP1 | CCAT | C | 0.034 | Non-frameshift insertion | 45/3/0 | SMAP1:NM_001281439:exon10:c.1223_1224insCAT:p.S408delinsSI |
| rs576516 | 71569090 | SMAP1 | C | T | 0.442 | Nonsyn. | 14/28/6 | SMAP1:NM_001281439:exon10:c.T1247C:p.M416T |
| rs200678168 | 71665982 | B3GAT2 | C | G | 0.018 | Nonsyn. | 43/5/0 | B3GAT2:NM_080742:exon1:c.C151G:p.R51G |
| rs112585190 | 71998778 | OGFRL1 | C | T | 0.118 | Nonsyn. | 36/11/1 | OGFRL1:NM_024576:exon1:c.T139C:p.S47P |
| rs72265796 | 72011579 | OGFRL1 | G | GAGA | 0.029 | Non-frameshift deletion | 46/2/0 | OGFRL1:NM_024576:exon7:c.1184_1186del:p.395_396del |
| rs767118962 | 72943520 | RIMS1 | T | C | Nonsyn | 47/1/0 | RIMS1:NM_001168407:exon2:c.C145T:p.R49W,RIMS1:NM_001168408:exon2:c.C145T:p.R49W,RIMS1:NM_001168410:exon2:c.C100T:p.R34W,RIMS1:NM_014989:exon7:c.C1723T:p.R575W | |
| rs764264653 | 72952068 | RIMS1 | G | A | Nonsyn | 47/1/0 | RIMS1:NM_001168407:exon5:c.A431G:p.E144G,RIMS1:NM_001168408:exon5:c.A431G:p.E144G,RIMS1:NM_001168409:exon5:c.A188G:p.E63G,RIMS1:NM_001168410:exon5:c.A386G:p.E129G,RIMS1:NM_014989:exon10:c.A2009G:p.E670G | |
| rs41265501 | 72984123 | RIMS1 | T | C | 0.025 | Nonsyn | 45/3/0 | RIMS1:NM_014989:exon23:c.C3470T:p.P1157L |
| Variant . | BP . | Gene . | Alt . | Ref . | MAF . | Effect . | Geno . | AA change . |
|---|---|---|---|---|---|---|---|---|
| rs6910140 | 70944257 | COL9A1 | C | T | 0.013 | Nonsyn | 47/1/0 | COL9A1:NM_078485:exon29:c.A1570G:p.M524V,COL9A1:NM_001851:exon35:c.A2299G:p.M767V |
| rs1135056 | 70961833 | COL9A1 | C | T | 0.407 | Nonsyn | 20/22/6 | COL9A1:NM_078485:exon22:c.A1133G:p.Q378R,COL9A1:NM_001851:exon28:c.A1862G:p.Q621R |
| rs592121 | 70984436 | COL9A1 | G | A | 0.367 | Nonsyn | 22/19/7 | COL9A1:NM_078485:exon5:c.T286C:p.S96P,COL9A1:NM_001851:exon11:c.T1015C:p.S339P |
| rs2747701 | 71238105 | FAM135A | G | A | 0.451 | Nonsyn | 19/17/12 | FAM135A:NM_001162529:exon14:c.A3725G:p.D1242G,FAM135A:NM_020819:exon15:c.A3086G:p.D1029G,FAM135A:NM_001105531:exon16:c.A3137G:p.D1046G |
| rs1048886 | 71289189 | C6orf57 | G | A | 0.206 | Nonsyn | 31/14/3 | C6orf57:NM_145267:exon2:c.A137G:p.Q46R |
| rs146446063 | 71298323 | C6orf57 | T | C | 0.007 | Nonsyn | 44/4/0 | C6orf57:NM_145267:exon3:c.C223T:p.P75S |
| rs112439957 | 71377781 | SMAP1 | A | C | 0.008 | Nonsyn | 44/4/0 | SMAP1:NM_001044305:exon1:c.C55A:p.L19I,SMAP1:NM_001281439:exon1:c.C55A:p.L19I,SMAP1:NM_021940:exon1:c.C55A:p.L19I |
| rs200042613 | 71378354 | SMAP1 | A | AC | 0.033 | Frameshift deletion | 45/3/0 | SMAP1:NM_001281440:exon1:c.24delC:p.H8fs |
| rs2273566 | 71546702 | SMAP1 | T | C | 0.084 | Nonsyn | 40/8/0 | SMAP1:NM_001281439:exon6:c.C554T:p.A185V,SMAP1:NM_021940:exon6:c.C554T:p.A185V,SMAP1:NM_001044305:exon7:c.C635T:p.A212V,SMAP1:NM_001281440:exon7:c.C605T:p.A202V |
| rs201201431 | 71569066 | SMAP1 | CCAT | C | 0.034 | Non-frameshift insertion | 45/3/0 | SMAP1:NM_001281439:exon10:c.1223_1224insCAT:p.S408delinsSI |
| rs576516 | 71569090 | SMAP1 | C | T | 0.442 | Nonsyn. | 14/28/6 | SMAP1:NM_001281439:exon10:c.T1247C:p.M416T |
| rs200678168 | 71665982 | B3GAT2 | C | G | 0.018 | Nonsyn. | 43/5/0 | B3GAT2:NM_080742:exon1:c.C151G:p.R51G |
| rs112585190 | 71998778 | OGFRL1 | C | T | 0.118 | Nonsyn. | 36/11/1 | OGFRL1:NM_024576:exon1:c.T139C:p.S47P |
| rs72265796 | 72011579 | OGFRL1 | G | GAGA | 0.029 | Non-frameshift deletion | 46/2/0 | OGFRL1:NM_024576:exon7:c.1184_1186del:p.395_396del |
| rs767118962 | 72943520 | RIMS1 | T | C | Nonsyn | 47/1/0 | RIMS1:NM_001168407:exon2:c.C145T:p.R49W,RIMS1:NM_001168408:exon2:c.C145T:p.R49W,RIMS1:NM_001168410:exon2:c.C100T:p.R34W,RIMS1:NM_014989:exon7:c.C1723T:p.R575W | |
| rs764264653 | 72952068 | RIMS1 | G | A | Nonsyn | 47/1/0 | RIMS1:NM_001168407:exon5:c.A431G:p.E144G,RIMS1:NM_001168408:exon5:c.A431G:p.E144G,RIMS1:NM_001168409:exon5:c.A188G:p.E63G,RIMS1:NM_001168410:exon5:c.A386G:p.E129G,RIMS1:NM_014989:exon10:c.A2009G:p.E670G | |
| rs41265501 | 72984123 | RIMS1 | T | C | 0.025 | Nonsyn | 45/3/0 | RIMS1:NM_014989:exon23:c.C3470T:p.P1157L |
AA, amino acid; BP: base pair position; Geno, genotype count (homozygous Ref/heterozygous/homozygous Alt).
Association of NGS target SNPs with pediatric VTE
A total of 10 361 variants (SNPs and indels) identified in the NGS target region were tested for association in 24 discordant siblings from stage I families using the SDT. Fourteen intronic RIMS1 variants and 34 intergenic variants near the genes B3GAT2, RIMS1, OGFRL1, and FAM135A showed a nominal P value <.05. Of 14 nonsynonymous coding SNPs and 3 indels discovered in the NGS (Table 2), 8 variants were forwarded to genotyping and association analysis in 189 father-mother–affected child trios from the initial stage I cohort (23 trios of this cohort were not available any longer because of a shortage of DNA sample material). Of these, rs72265796 was removed from further analyses because of calling errors. Three SNPs curtly missed statistical significance in the transmission-disequilibrium test with rs112439957 (P = .08326, SMAP1), rs767118962 (P = .08326, RIMS1), and rs41265501 (P = .05778, RIMS1) likely resulting from power restrictions (supplemental Table 4). Similar findings were observed when using the FBAT-weighted rare variant statistic for variants at a minor allele frequency (MAF) < 0.03. The best composite P value was observed for the rare variants residing within the SMAP1 gene (P = .0660).
Discussion
In the past several years, numerous GWAS have confirmed established risk factors, such as FV, FGG, and the ABO locus, and yielded novel susceptibility genes for VTE in adults.7-11 Here we present the first GWAS on VTE in children using the Infinium 660W SNP array and a large collection of affected nuclear families comprising 212 parent-child trios. From the loci previously implicated through GWAS in adults, we were able to confirm SNPs located in the ABO locus, F11 and CYP4V2, in addition to the “classical” genetic risk factors, prothrombin, FV, FGA, and FGG, previously published as part of candidate gene approaches.1-6 Our data further strengthen the role of these genetic susceptibility factors in the pathogenesis of VTE in both children and adults.
In our GWAS analysis, the most significant association was found in a genomic region on chromosome 6q13 comprising the SMAP1 and B3GAT2 genes, and a Rab-3 interacting molecule (RIMS1) downstream of the main association signal, with confident P values (P < 10−5), implicating 3 novel loci in the pathogenesis of pediatric VTE. This initial association was validated in an independent study sample of children with TS, which served as a pseudo-replication. A second replication attempt in a sample of adult patients with VTE and controls coming from the PopGen biobank13 showed an association of 2 SNPs in the SMAP1/B3GAT2 region at a nominal P value <.05. However, after correction for multiple testing, this association missed the threshold of significance and therefore needs to be viewed with caution. This may be due to either limited sample size of this replication sample, and thus power limitations, or the phenotype window, meaning that the association observed in our pediatric samples may not be relevant for studies in adults. This also points toward a limitation of our study. Because no other pediatric VTE cohorts were available for validation, both replication cohorts, which differ slightly with respect to clinical phenotypes, were the only option for pseudo-replication. Stage II probands with pediatric TS include thromboses of cardiac origin that are more frequently provoked by mechanical stress than venous thromboses from paradoxical origin. VTE location differs slightly between pediatric venous thromboses of stage I and stage III, with more emphasis on leg thromboses in adult patients (supplemental Table 2). Furthermore, provoked VTE was found more frequently in adults (supplemental Table 2). Nevertheless, we believe that our 2 independent replication studies give valuable indication for our association results to be true findings.
Because rare, coding, or other functionally relevant variants residing in 3′ untranslated regions, 5′ untranslated regions, or splice junction sites may explain the observed association and pinpoint the true causative gene mutation(s), we used targeted resequencing of 24 discordant sibling pairs, a subset of the initial GWAS cohort, in an attempt to identify such functionally relevant variants. In total, 14 nonsynonymous SNPs and 3 indels were identified, 7 of which are predicted to be damaging or deleterious based on Protein Variation Effect Analyzer and sorting intolerant from tolerant scores. Interestingly, we found 3 nonsynonymous coding SNPs and 2 frameshift deletions/insertions in SMAP1 and 3 nonsynonymous SNPs in RIMS1, whereas only 1 nonsynonymous SNP was found in B3GAT2, which is predicted to be neutral. This renders RIMS1 and SMAP1 attractive candidate genes for VTE, particularly in light of the observation that 3 nonsynonymous mutations only curtly miss significance when tested for association in the initial stage I cohort with 189 trios, despite the obvious power limitations of rare variant detection algorithms in complex diseases.28
There are only a few reports on the function of SMAP1 and B3GAT2, particularly in relation to a potential role in hemostasis. SMAP1, a GTPase-activating protein specific for adenosine 5′-diphosphate-ribosylation factor 6 (ARF6), is a small GTPase acting on membrane trafficking and actin remodeling. In neutrophils and macrophages, ARF6 is known to regulate a variety of functions (eg, phagocytosis, β2 integrin-dependent adhesion, Toll-like receptor signaling)29 as well as platelet spreading and clot retraction.30 As shown in mouse models, SMAP1 messenger RNA is expressed in the erythroid lineage and other hematopoietic lineages, including progenitors.31 A large proportion of SMAP1 knockout mice (SMAP1−/−) developed hematological disorders, such as a decrease in platelet number, pointing toward an important role of SMAP1 in the hematopoiesis in general.31
Comparably little is known about the function of B3GAT2. Thus far, the gene has been described in various mammalian species and encodes for a transmembrane protein belonging to the glucuronyltransferase family.32,33 B3GAT2 is primarily expressed in neuronal tissues and is involved in the initial steps of proteoglycan synthesis, including synthesis of the HNK-1 carbohydrate epitope.32 Previously, a GWAS reported SNPs in B3GAT2 to be significantly associated with distribution of fetal hemoglobin-containing erythrocytes (F cells) at a P = 9 × 10−7 in 440 individuals with sickle cell disease (SCD) of African ancestry.34 SCD is associated with a chronically activated coagulation system, as evidenced by increased markers of thrombin generation, decreased levels of anticoagulant proteins, and an activation of the fibrinolytic system.35 Interestingly, Austin and colleagues36 reported an increased risk for VTE (OR, 1.8) and pulmonary embolism (OR, 3.9) in 515 black patients carrying the sickle cell trait. Although the sickle cell trait is not prevalent in Caucasians, these studies implicate B3GAT2 as an important molecule in hemostasis, particularly in the transition from the fetal to the adult hemostatic system after birth. This is apparently of relevance for pediatric VTE because a significant number of pediatric incidences occur in neonates,37 but leaves room for speculation about its functional relevance in adults.
RIMS1, which is located ∼1.2 Mb downstream from the peak association signal and located on a different haplotype block than SMAP1/B3GAT2, appears to contribute to a second, though less pronounced, association peak in this genomic region. However, because we detected 2 nonsynonymous SNPs, which were predicted to be detrimental and curtly missed significance in the full cohort, we consider this gene as another positional candidate for VTE. RIMS1 is a member of the RIM family of active zone proteins, a synaptic vesicle protein, and regulates synaptic vesicle exocytosis.38,39 RIMS1 is also expressed in pancreatic β cells, where it is involved in insulin release by exocytosis.40 Various components of the neuronal vesicle release machinery are also present in other cell types that contain secretory granules (eg, platelets,41 neutrophils42 ) and one might speculate that RIMS1 is an integral component of the exocytosis scaffolds in these cells. The Gene Expression Omnibus database has 3 entries on RIMS1 being expressed in platelets from SCD patients. However, whether or not RIMS1 is expressed in normal or prothrombotic platelets (or other compartments of relevance for hemostasis) remains to be elucidated in functional studies.
Taken together all 3 genes—SMAP1, B3GAT2, and RIMS1—are plausible positional candidate genes contributing to the association signal observed in our GWAS based on our NGS analysis as well as functional plausibility. The question, whether 1 or several genes residing in this region contribute to VTE cannot be solved in a genetic association study. The dissection of the genetic causes in a complex genomic region such as this, with up to 13 positional candidate genes, warrants detailed functional workup that exceeds the scope of the current study.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all hospitals and physicians contributing cases and controls to this study and all families who participated. The authors also thank Rory Koenen from Maastricht University for careful reading and valuable comments on the manuscript. Furthermore, the authors acknowledge the excellent technical assistance of Marianne Jansen-Rust, Elvira Barg, and Gerrit Randau from the Core Facility Genomics of the Medical Faculty Münster.
The pediatric stroke study (replication cohort) was supported by grants from the Förderverein “Schlaganfall und Thrombosen im Kindesalter e.V.” and Interdisziplinäres Zentrum für Klinische Forschung (CRA01-09), University of Muenster, Germany. The sponsors had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, preparation, review, or approval of the manuscript, and decision to submit the manuscript for publication.
Authorship
Contribution: M.S. and U.N.-G. were responsible for the study concept and design; U.N.-G. acquired the clinical data; F.R., A.A., M.H., and M.R. undertook the genome-wide association studies and replication study analysis; A.H. and A.B. analyzed the next-generation sequencing data; C.H., A.K., V.L., R.M., and U.N.-G. were responsible for patient recruitment (pediatric and adult); A.F. and W.L. provided DNA samples and demographic data from PopGen; M.S. drafted the manuscript; genotyping was performed in the Core Facility Genomics of the Medical Faculty Münster by A.W.; and all authors contributed to the interpretation of data and critically revised the report for important intellectual content.
Conflict-of-interest disclosure: A.K. has received refunding of travel costs from Baxter, Novo Nordisk, and Commonwealth Serum Laboratories Behring unrelated to this study. The remaining authors declare no competing financial interests.
Correspondence: Monika Stoll, Institute for Human Genetics, Genetic Epidemiology, Westfälische Wilhelms University of Muenster, Albert-Schweitzer-Campus 1, D3, 48149 Muenster, Germany; e-mail: mstoll@uni-muenster.de.
References
Author notes
U.N.-G. and M.S. are joint senior authors.