Key Points
Inhibition of SPHK1 in human AML cells induces MCL1 degradation and caspase-dependent cell death.
SPHK1 inhibitors reduce leukemic burden and prolong survival in orthotopic patient-derived xenografts of AML.
Abstract
Acute myeloid leukemia (AML) is an aggressive malignancy where despite improvements in conventional chemotherapy and bone marrow transplantation, overall survival remains poor. Sphingosine kinase 1 (SPHK1) generates the bioactive lipid sphingosine 1-phosphate (S1P) and has established roles in tumor initiation, progression, and chemotherapy resistance in a wide range of cancers. The role and targeting of SPHK1 in primary AML, however, has not been previously investigated. Here we show that SPHK1 is overexpressed and constitutively activated in primary AML patient blasts but not in normal mononuclear cells. Subsequent targeting of SPHK1 induced caspase-dependent cell death in AML cell lines, primary AML patient blasts, and isolated AML patient leukemic progenitor/stem cells, with negligible effects on normal bone marrow CD34+ progenitors from healthy donors. Furthermore, administration of SPHK1 inhibitors to orthotopic AML patient–derived xenografts reduced tumor burden and prolonged overall survival without affecting murine hematopoiesis. SPHK1 inhibition was associated with reduced survival signaling from S1P receptor 2, resulting in selective downregulation of the prosurvival protein MCL1. Subsequent analysis showed that the combination of BH3 mimetics with either SPHK1 inhibition or S1P receptor 2 antagonism triggered synergistic AML cell death. These results support the notion that SPHK1 is a bona fide therapeutic target for the treatment of AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematological malignancy presenting as an accumulation of immature myeloid cells in the bone marrow and peripheral blood. Despite improvements in our understanding of the molecular evolution of this disease, the overall survival of young adults (<60 years) is <30%.1 New disease targeting modalities such as kinase inhibitors, epigenetic modifiers, and monoclonal antibodies have recently been developed; however, results from clinical trials have been disappointing,2 and currently, no targeted therapies are approved for routine clinical use.
Sphingosine kinase 1 (SPHK1) generates the bioactive lipid sphingosine 1-phosphate (S1P) that promotes several of the biological hallmarks of cancer, including cell survival and proliferation through its action as either a ligand for a family of 5 S1P-specific G protein–coupled receptors (S1PR1-5) or an intracellular second messenger.3,4 Many studies have reported that high SPHK1 expression in solid tumors is frequently associated with increased disease progression, chemoresistance, and poor prognosis.5 Indeed, targeting of SPHK1 with either small-molecule inhibitors or via genetic ablation has proved efficacious in blocking tumor progression in mouse models of diverse human solid cancers.6-14 Several studies have recently implicated a role for SPHK1 in leukemogenesis.15 For example, SPHK1 inhibition has been shown to sensitize leukemic cells to chemotherapy,16,17 directly induce cell death in HL-60 AML cells,18,19 and reduce the growth of subcutaneous U937 AML cell line xenografts in mice,20,21 but the mechanism of action and the efficacy in primary AML have not been studied.
Here, we examined the role and targeting of SPHK1 in primary AML patient cells including those of the stem and progenitor compartment. We found that primary AML blasts, as well as isolated CD34+/CD38−/CD123+ leukemic stem and progenitor cells (LSPCs), are sensitive to SPHK1 inhibition both in vitro and in orthotopic xenografts in mice. Mechanistically, we found that AML cell apoptosis induced by SPHK1 inhibition was attributable to a loss of the prosurvival protein MCL1 caused by decreased signaling through S1P receptor 2. Because MCL1 has emerged as a critical target in many different cancers, our studies suggest that targeting SPHK1 to block MCL1 expression may have clinical utility in AML and other malignancies that have high dependency on MCL1.
Methods
Cell lines and primary AML samples
Microarray data of SPHK1 messenger RNA (mRNA) levels from fluorescence-activated cell sorter (FACS)–purified hematopoietic stem cells (HSCs; Lin−/CD34+/CD38−/CD90+/CD45RA−) and AML cells from various cytogenetic subgroups were obtained from BloodSpot using the BloodPool data set, AML samples with normal cells (http://servers.binf.ku.dk/bloodspot/?gene;).22 AML RNA sequencing (RNA-Seq) data were obtained from The Cancer Genome Atlas (TCGA; RNA Expression Level 3 Data Archives [DCC] IlluminaGA RNASeq at https://tcga-data.nci.nih.gov/docs/publications/laml_2012/). Normal bone marrow (NBM) RNA-Seq data were obtained from the Human Protein Atlas file (E-MTAB-1733).23 Significance was assessed by Student t test. The AML cell lines ME-1, MOLM-13, MV4-11, and THP-1 cells were cultured in RPMI supplemented with 10% fetal calf serum (FCS; HyClone Thermo Scientific). The factor-dependent cells TF-1 and UT-7 were grown as previously described.24 Cell line authentication was confirmed by short tandem repeats profiling.
Mononuclear cells (MNC) from diagnostic bone marrow or apheresis product samples were isolated by Ficoll-Hypaque density-gradient centrifugation and resuspended in Iscove modified Dulbecco medium containing 10% FCS.25 FACS purification of primary human CD34+/CD38−/CD123+ LSPCs was performed as previously described.26 Cell lysates were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotted using the anti-SPHK1 (ECM Biosciences), anti-Ser225 SPHK1 (ECM Biosciences), anti-SPHK2 (Proteintech), or anti-β-actin (Merck Millipore). All other antibodies were purchased from Cell Signaling Technology. SPHK1 activity assays were performed as previously described.27
Mutation analyses of AML biopsies
Mutational analysis of FLT3-ITD (internal tandem duplication) and NPM1 (nucleophosmin) in AML biopsies was performed as previously described.28-30 Mutations affecting KIT D816, DMNT3A R882, FLT3-TKD D835/I836, IDH1 R132, IDH2 R140/R172, JAK1 T478/V623, JAK2 V617, KRAS G12/13, MPL W515, NPM1 W288, and NRAS G12/13 and Q61 were detected using a multiplexed matrix-assisted laser desorption/ionization time-of-flight genotyping approach (Sequenom MassARRAY Compact System) as previously described.30
Constructs and mRNA analysis
Details of construct generation, RNA-Seq of MP-A08–treated cells, bioinformatics, and quantitative reverse transcription polymerase chain reaction (RT-PCR) are presented in the supplemental Methods (available on the Blood Web site).
Cell survival and colony assays
Cell survival was determined by annexin V–fluorescein isothiocyanate (Roche) negativity and/or propidium iodide (PI) exclusion using a FACS (Gallios, Beckman Coulter).31 Primary AML blasts, LSPCs, and NBM-derived CD34+ cells were cultured in Iscove modified Dulbecco medium supplemented with 0.5% FCS. Caspase-3 activity was assessed by substrate cleavage of NucViewTM488 (Biotium). Colony assays from bone marrow cells where performed in triplicate with H4230 Stem Cell Technology methyl cellulose medium supplemented with 2 ng/mL interleukin 3 and granulocyte-macrophage colony-stimulating factor as previously described.31
Xenotransplantation
Transplantation of human cells into female NOD/SCID (NOD.CB17-Prkdcscid/J) mice was performed as previously described.30 Briefly, mice were sublethally irradiated (275 cGy) 24 hours prior to IV injection of 5–10 × 106 primary human AML cells via tail vein. Engraftment levels were quantified by flow cytometry and expressed as the percentage of human CD45+ (hCD45+) cells to total hCD45+ and mouse CD45+ (mCD45+) cells in the bone marrow.32 MP-A08 (100 mg/kg) or vehicle (70% [vol/vol] PEG400) was administered by intraperitoneal (IP) injection 5 to 6 times a week for 2 to 5 weeks. Immunohistochemistry on mouse bone marrow was performed as previously described33 with human mitochondrial antibodies (Abcam). An event was defined to occur when the mice exhibited signs of leukemia related morbidity and the bone marrow exhibited >25% hCD45 staining.
Study approval
All studies were approved by the SA Pathology Institutional Biosafety Committee. Animal studies were performed under the institutional guidelines approved by the SA Pathology/Central Adelaide Local Health Network Animal Ethics Committee. Human samples were obtained from patients diagnosed with AML after informed consent according to institutional guidelines, and studies were approved by the Royal Adelaide Hospital Human Research Ethics Committee.
Results
SPHK1 is overexpressed in AML
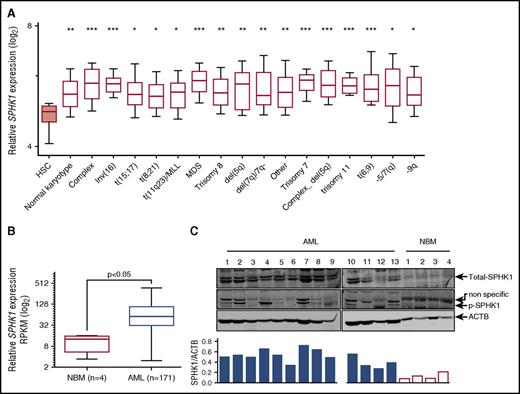
To examine SPHK1 as a potential target in AML, we first assessed SPHK1 mRNA expression levels in AML cells derived from patients of diverse disease subtypes (n = 1933) using BloodSpot,22 an integrated microarray data portal for the analysis of FACS-sorted healthy and malignant hematopoietic cells. We found mean SPHK1 mRNA expression was higher in all cytogenetic subclasses of AML compared with normal HSCs (Lin−/CD34+/CD38−/CD90+/CD45RA−) (Figure 1A). This overexpression was particularly high in complex karyotype AML. By comparison, the other sphingosine kinase isoform, SPHK2, was not significantly overexpressed across all of the cytogenetic subclasses (supplemental Figure 1). We next examined SPHK1 mRNA expression levels in AML cells (n = 171) using TCGA. We found mean SPHK1 mRNA expression was sixfold higher in AML compared with NBM (Figure 1B). This overexpression was not associated with any specific cytogenetic abnormalities risk or mutational status of IDH1, NPM1, FLT3, or RAS (supplemental Figure 2). We also examined SPHK1 protein expression in MNCs obtained from AML patients. Two SPHK1 splice isoforms, SPHK1a and SPHK1c, were expressed in all samples examined (Figure 1C; n = 13), consistent with previous reports in AML cell lines.19,20 Importantly, SPHK1 protein was below detection limits in MNCs obtained from the bone marrow of healthy volunteers. We have previously shown that activation of SPHK1 via phosphorylation of Ser225 by extracellular signal-regulated kinases 1/2 (ERK1/2) promotes oncogenic signaling by this enzyme.34,35 Consistent with this, we found that levels of Ser225 phosphorylation of SPHK1 were elevated in AML patient blasts compared with MNCs from healthy volunteers (Figure 1C) signifying elevated SPHK1 signaling.
SPHK1 is overexpressed in AML. (A) Microarray analysis of SPHK1 mRNA levels from FACS-purified HSCs (Lin−/CD34+/CD38−/CD90+/CD45RA−) and AML cells from various cytogenetic subgroups obtained from BloodSpot (median, 25 to 75 percentiles boxed, and 10 to 90 percentiles shown with bars).22 Significance was assessed by Student t test (*P < .05; **P < .01; ***P < .001) for HSCs compared with all AML subtypes shown. (B) Normalized RNA-Seq reads (reads per kilobase per million mapped reads; RPKM) for SPHK1 from primary AML patient samples (n = 171) and normal bone marrow samples (n = 4) were obtained from TCGA and the Human Protein Atlas, respectively. Significance was assessed by Student t test (P < .05). (C) Primary AML blasts or normal bone marrow MNCs where lysed and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by western blot analysis of total SPHK1 and phospho-Ser225 SPHK1. Western blot results were quantified by laser densitometry and expressed as the ratio of SPHK1/ACTB.
SPHK1 is overexpressed in AML. (A) Microarray analysis of SPHK1 mRNA levels from FACS-purified HSCs (Lin−/CD34+/CD38−/CD90+/CD45RA−) and AML cells from various cytogenetic subgroups obtained from BloodSpot (median, 25 to 75 percentiles boxed, and 10 to 90 percentiles shown with bars).22 Significance was assessed by Student t test (*P < .05; **P < .01; ***P < .001) for HSCs compared with all AML subtypes shown. (B) Normalized RNA-Seq reads (reads per kilobase per million mapped reads; RPKM) for SPHK1 from primary AML patient samples (n = 171) and normal bone marrow samples (n = 4) were obtained from TCGA and the Human Protein Atlas, respectively. Significance was assessed by Student t test (P < .05). (C) Primary AML blasts or normal bone marrow MNCs where lysed and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by western blot analysis of total SPHK1 and phospho-Ser225 SPHK1. Western blot results were quantified by laser densitometry and expressed as the ratio of SPHK1/ACTB.
Targeting SPHK1 induces AML cell death
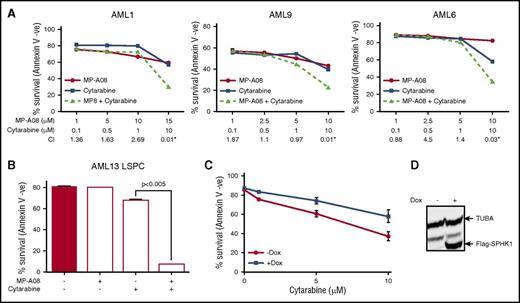
To assess the efficacy of targeting SPHK1 in AML, we initially screened a panel of AML cell lines with MP-A08, a highly selective sphingosine kinase inhibitor with a Ki for SPHK1 of 27 μM.33 All 6 AML cell lines exhibited sensitivity to SPHK1 inhibition (Figure 2A) with 50% inhibitory concentration (IC50) values ranging between 6.8 and 25.8 µM. To ascertain the clinical potential of targeting SPHK1 in AML, a panel of 20 AML patient samples was screened to determine the IC50 in AML blasts (Figure 2B; Table 1). Samples were profiled for mutations in KIT, DNMT3A, FLT3, IDH1, IDH2, JAK1, JAK2, KRAS, MPL, NPM1, and NRAS (Table 1). The majority of AML cases were susceptible to MP-A08 with IC50 values ranging between 6.4 and 18.9 µM. Comparable sensitivities were observed when AML blasts were treated with SKI-II, a sphingosine-competitive sphingosine kinase inhibitor14 (Table 1). This in vitro activity did not cluster with any particular mutation group. Furthermore, MP-A08 was effective at blocking leukemic colony-forming units from primary AML samples (supplemental Figure 3). In contrast to the susceptibility of AML cells, normal CD34+ bone marrow progenitors were considerably more resistant to SPHK1 inhibition (Figure 2C), and indeed, minimal effects were seen on these cells with MP-A08 up to 30 μM, suggesting a potential therapeutic window for the selective targeting of AML cells. Next, we assessed the efficacy of targeting SPHK1 in LSPCs that are reported to drive relapse and have intrinsic resistance to chemotherapy.36 FACS-purified CD34+/CD38−/CD123+ LSPCs from AML patients showed similar sensitivity to SPHK1 inhibition by MP-A08 as the total blasts (Figure 2D; Table 1).
Targeting SPHK1 induces AML cell death. AML cell lines (A), primary AML blasts (B), or normal bone marrow–derived CD34+ cells (C) were plated in the indicated concentrations of MP-A08, and cell survival was determined by annexin V staining. (D) FACS-purified primary human CD34+/CD38−/CD123+ LSPCs were treated with a dose response of MP-A08 for 24 hours, and cell survival was determined by annexin V staining. Results are mean ± range. MV4-11 cells were transduced with doxycycline-inducible short hairpin RNA (shRNA) constructs to knockdown SPHK1 and a nontargeting control shRNA. Cells were incubated with 1 μg/mL doxycycline (Dox) and 48 hours postinduction quantitative RT-PCR (mean ± standard deviation, n = 3) (E), SPHK1 activity assays (mean ± range) (F), immunoblot analysis of whole cell lysates were performed and quantified by laser densitometry (ratio of SPHK1/ACTB) (G), and analysis of cell survival was determined by annexin V staining (mean ± standard error of the mean [SEM], n = 3) (H). Significance was assessed by Student t test.
Targeting SPHK1 induces AML cell death. AML cell lines (A), primary AML blasts (B), or normal bone marrow–derived CD34+ cells (C) were plated in the indicated concentrations of MP-A08, and cell survival was determined by annexin V staining. (D) FACS-purified primary human CD34+/CD38−/CD123+ LSPCs were treated with a dose response of MP-A08 for 24 hours, and cell survival was determined by annexin V staining. Results are mean ± range. MV4-11 cells were transduced with doxycycline-inducible short hairpin RNA (shRNA) constructs to knockdown SPHK1 and a nontargeting control shRNA. Cells were incubated with 1 μg/mL doxycycline (Dox) and 48 hours postinduction quantitative RT-PCR (mean ± standard deviation, n = 3) (E), SPHK1 activity assays (mean ± range) (F), immunoblot analysis of whole cell lysates were performed and quantified by laser densitometry (ratio of SPHK1/ACTB) (G), and analysis of cell survival was determined by annexin V staining (mean ± standard error of the mean [SEM], n = 3) (H). Significance was assessed by Student t test.
Clinical features of AML patient samples and sensitivity to MP-A08
| Patient ID . | Age/sex . | Cytogenetics . | FAB . | Marrow blasts (%) . | WCC × 109/L . | Mutational status . | MP-A08 IC50 (µM) . | SKI-II IC50 (µM) . | MP-A08 LSPC IC50 (µM) . |
|---|---|---|---|---|---|---|---|---|---|
| AML1 | 56/M | Normal | M4 | 75 | 132 | FLT3-ITD, IDH1 R132H | 8.8 | 9.8 | 8.1 |
| AML2 | 32/M | t(9;11), 11q23 | M5 | 90 | 55 | None | N/D | N/D | N/D |
| AML3 | 62/M | +1,de(1;6) | N/D | 55 | 33 | None* | 12.1 | N/D | N/D |
| AML4 | 77/M | Normal | M1 | 96 | 46 | FLT3-ITD, NPM1, IDH2 R140Q, DNMT3A R882H | N/D | N/D | N/D |
| AML5 | 72/F | −5q | M1 | 90 | 104 | FLT3-TKD* | N/D | N/D | N/D |
| AML6 | 64/F | del(16q) | M1 | 100 | 111 | None | 14.8 | 11.7 | N/D |
| AML7 | 62/F | t(2;11), 11q23 | M4 | 97 | 131 | None* | 10.1 | 6.2 | 11.2 |
| AML8 | 44/F | t(9;11), 11q23 | N/D | 82 | 221 | N/D | 18.9 | 10.6 | N/D |
| AML9 | 39/F | Normal | M1 | 91 | 122 | FLT3-ITD, NPM1, IDH2 R140Q | N/D | N/D | N/D |
| AML10 | 69/F | Normal | M1 | 98 | 107 | FLT3-ITD, IDH2 R140Q | 11.9 | N/D | N/D |
| AML11 | 55/F | Normal | M4 | 39 | 67 | FLT3-ITD | N/D | N/D | N/D |
| AML12 | 63/M | t(8;21) | M2 | 65 | 35 | FLT3-ITD | N/D | N/D | N/D |
| AML13 | 75/M | +8 | M5 | 87 | 227 | FLT3-TKD D835Y, IDH2 R140Q | 14.7 | 10.0 | 13.1 |
| AML14 | 20/M | Normal | M4 | 79 | 94 | FLT3-ITD | 13.6 | 7.8 | 13.1 |
| AML15 | 60/F | −9q | M2 | 47 | 22 | None | 8.5 | 6.1 | N/D |
| AML16 | 51/M | Inv 16 | M4 | 31 | 117 | FLT3-ITD | 12.7 | 4.6 | N/D |
| AML17 | 72/F | Normal | M4 | 96 | 91 | FLT3-ITD, NPM1* | 6.4 | 6.6 | N/D |
| AML18 | 56/M | Complex | M2 | 24 | 6.3 | None | 16.8 | N/D | N/D |
| AML19 | 59/F | Normal | M2 | 63 | 119 | FLT3-ITD, NPM1, DNMT3A R882H | 16.0 | N/D | N/D |
| AML20 | 78/M | Normal | M2 | 55 | 166 | DNMT3A R882H | 11.5 | N/D | N/D |
| AML21 | 63/M | Normal | M1 | 96 | 140 | NPM1* | 11.7 | N/D | N/D |
| AML22 | 59/F | Normal | N/D | 87 | 385 | N/D | 10.1 | N/D | N/D |
| AML23 | 37/F | Normal | M4 | NA | 3.9 | FLT3-ITD, NPM1 | 12.3 | N/D | N/D |
| AML24 | 61/F | Normal | M4 | 71 | 19 | FLT3-ITD, NPM1, IDH2 R140Q | 12.5 | N/D | N/D |
| AML25 | 56/F | Normal | M1 | 89 | 57 | None* | 11.0 | N/D | N/D |
| AML26 | 67/M | −9q | N/D | 53 | 2.96 | N/D | 12.4 | N/D | N/D |
| AML27 | 47/F | −9q | M2 | 21 | 2.8 | None | N/D | N/D | N/D |
| AML28 | 86/M | +8 | M4 | 46 | 181 | FLT3-ITD | N/D | N/D | N/D |
| AML29 | 81/M | –Y,t(10;11), 11q23 | M5 | 92 | 178 | None* | N/D | N/D | N/D |
| AML30 | 23/M | inv16 | M4 | 88 | 60 | FLT3-ITD* | N/D | N/D | N/D |
| Patient ID . | Age/sex . | Cytogenetics . | FAB . | Marrow blasts (%) . | WCC × 109/L . | Mutational status . | MP-A08 IC50 (µM) . | SKI-II IC50 (µM) . | MP-A08 LSPC IC50 (µM) . |
|---|---|---|---|---|---|---|---|---|---|
| AML1 | 56/M | Normal | M4 | 75 | 132 | FLT3-ITD, IDH1 R132H | 8.8 | 9.8 | 8.1 |
| AML2 | 32/M | t(9;11), 11q23 | M5 | 90 | 55 | None | N/D | N/D | N/D |
| AML3 | 62/M | +1,de(1;6) | N/D | 55 | 33 | None* | 12.1 | N/D | N/D |
| AML4 | 77/M | Normal | M1 | 96 | 46 | FLT3-ITD, NPM1, IDH2 R140Q, DNMT3A R882H | N/D | N/D | N/D |
| AML5 | 72/F | −5q | M1 | 90 | 104 | FLT3-TKD* | N/D | N/D | N/D |
| AML6 | 64/F | del(16q) | M1 | 100 | 111 | None | 14.8 | 11.7 | N/D |
| AML7 | 62/F | t(2;11), 11q23 | M4 | 97 | 131 | None* | 10.1 | 6.2 | 11.2 |
| AML8 | 44/F | t(9;11), 11q23 | N/D | 82 | 221 | N/D | 18.9 | 10.6 | N/D |
| AML9 | 39/F | Normal | M1 | 91 | 122 | FLT3-ITD, NPM1, IDH2 R140Q | N/D | N/D | N/D |
| AML10 | 69/F | Normal | M1 | 98 | 107 | FLT3-ITD, IDH2 R140Q | 11.9 | N/D | N/D |
| AML11 | 55/F | Normal | M4 | 39 | 67 | FLT3-ITD | N/D | N/D | N/D |
| AML12 | 63/M | t(8;21) | M2 | 65 | 35 | FLT3-ITD | N/D | N/D | N/D |
| AML13 | 75/M | +8 | M5 | 87 | 227 | FLT3-TKD D835Y, IDH2 R140Q | 14.7 | 10.0 | 13.1 |
| AML14 | 20/M | Normal | M4 | 79 | 94 | FLT3-ITD | 13.6 | 7.8 | 13.1 |
| AML15 | 60/F | −9q | M2 | 47 | 22 | None | 8.5 | 6.1 | N/D |
| AML16 | 51/M | Inv 16 | M4 | 31 | 117 | FLT3-ITD | 12.7 | 4.6 | N/D |
| AML17 | 72/F | Normal | M4 | 96 | 91 | FLT3-ITD, NPM1* | 6.4 | 6.6 | N/D |
| AML18 | 56/M | Complex | M2 | 24 | 6.3 | None | 16.8 | N/D | N/D |
| AML19 | 59/F | Normal | M2 | 63 | 119 | FLT3-ITD, NPM1, DNMT3A R882H | 16.0 | N/D | N/D |
| AML20 | 78/M | Normal | M2 | 55 | 166 | DNMT3A R882H | 11.5 | N/D | N/D |
| AML21 | 63/M | Normal | M1 | 96 | 140 | NPM1* | 11.7 | N/D | N/D |
| AML22 | 59/F | Normal | N/D | 87 | 385 | N/D | 10.1 | N/D | N/D |
| AML23 | 37/F | Normal | M4 | NA | 3.9 | FLT3-ITD, NPM1 | 12.3 | N/D | N/D |
| AML24 | 61/F | Normal | M4 | 71 | 19 | FLT3-ITD, NPM1, IDH2 R140Q | 12.5 | N/D | N/D |
| AML25 | 56/F | Normal | M1 | 89 | 57 | None* | 11.0 | N/D | N/D |
| AML26 | 67/M | −9q | N/D | 53 | 2.96 | N/D | 12.4 | N/D | N/D |
| AML27 | 47/F | −9q | M2 | 21 | 2.8 | None | N/D | N/D | N/D |
| AML28 | 86/M | +8 | M4 | 46 | 181 | FLT3-ITD | N/D | N/D | N/D |
| AML29 | 81/M | –Y,t(10;11), 11q23 | M5 | 92 | 178 | None* | N/D | N/D | N/D |
| AML30 | 23/M | inv16 | M4 | 88 | 60 | FLT3-ITD* | N/D | N/D | N/D |
FAB, French-American-British classification; N/D, not determined; WCC, white blood cell count.
Only FLT3-ITD, FLT3-TKD, and NPM1 screened.
To validate the inhibitor studies, we performed doxycycline-inducible shRNA-mediated SPHK1 knockdown, which resulted in a >50% reduction in SPHK1 mRNA, activity, and protein levels at 48 hours postinduction compared with the nontargeting control cells (Figure 2E-G). SPHK1 knockdown resulted in a 60% reduction in cell survival compared with the nontargeting control cells (Figure 2H). Similar studies with shRNA-mediated knockdown of SPHK2 showed no effect on AML cell survival (supplemental Figure 4). Collectively, these data suggest that targeting SPHK1 may have utility against diverse disease subtypes including those with poor prognosis and prone to chemotherapeutic failure.
Targeting SPHK1 chemosensitizes AML cells to cytarabine
We next examined the chemosensitizing effects of SPHK1 inhibition by MP-A08. Titration of MP-A08 with cytarabine, a pyrimidine nucleoside analog widely employed in standard induction chemotherapy for AML,37 resulted in synergistic cell death in both primary AML blasts (Figure 3A) and LSPCs (Figure 3B) (CI <1.0 and fractional product method with values <−0.1).38,39 To validate these findings, we employed a doxycycline-inducible lentiviral system to manipulate SPHK1 expression in MV4-11 cells. We found that doxycycline-induced SPHK1 overexpression resulted in enhanced chemotherapeutic resistance to cytarabine (Figure 3C). These results support the findings that targeting SPHK1 sensitizes AML cells to standard induction chemotherapeutics.
SPHK1 mediates chemotherapeutic resistance in AML. (A) Primary AML blasts where treated with subcytotoxic concentrations of MP-A08 (●), cytarabine (▪), or a combination of both (▲), and cell survival was quantified at 24 to 48 hours by annexin V staining. Synergism was assessed by the Chou-Talalay combination index (CI) using CalcuSyn software, where CI values <1 indicate synergism, or the fractional product method, where values <−0.1 indicate synergism (AML1 = −0.18, AML9 = −0.13, AML6 = −0.27). (B) FACS-purified primary human CD34+/CD38−/CD123+ LSPCs were treated with subcytotoxic concentrations of MP-A08, cytarabine, or a combination of both for 48 hours, and cell survival was determined by annexin V staining. (C) MV4-11 cells were transduced with a doxycycline-inducible lentiviral vector to overexpress FLAG-tagged SPHK1. Twenty-four hours after induction, cells were treated with increasing doses of MP-A08 for 48 hours, and cell survival was determined by annexin V staining. Results are mean ± range. (D) Immunoblot analysis of whole cell lysates with anti-FLAG confirmed doxycycline-inducible expression.
SPHK1 mediates chemotherapeutic resistance in AML. (A) Primary AML blasts where treated with subcytotoxic concentrations of MP-A08 (●), cytarabine (▪), or a combination of both (▲), and cell survival was quantified at 24 to 48 hours by annexin V staining. Synergism was assessed by the Chou-Talalay combination index (CI) using CalcuSyn software, where CI values <1 indicate synergism, or the fractional product method, where values <−0.1 indicate synergism (AML1 = −0.18, AML9 = −0.13, AML6 = −0.27). (B) FACS-purified primary human CD34+/CD38−/CD123+ LSPCs were treated with subcytotoxic concentrations of MP-A08, cytarabine, or a combination of both for 48 hours, and cell survival was determined by annexin V staining. (C) MV4-11 cells were transduced with a doxycycline-inducible lentiviral vector to overexpress FLAG-tagged SPHK1. Twenty-four hours after induction, cells were treated with increasing doses of MP-A08 for 48 hours, and cell survival was determined by annexin V staining. Results are mean ± range. (D) Immunoblot analysis of whole cell lysates with anti-FLAG confirmed doxycycline-inducible expression.
Targeting SPHK1 reduces leukemic burden and prolongs survival in vivo
Prior to treating established disease in an orthotopic mouse xenotransplantation model of human AML, we first examined potential adverse effects of MP-A08 on immunophenotypically defined hematopoietic bone marrow lineages. We previously demonstrated that MP-A08 is well tolerated in NOD/SCID mice with no adverse effects seen in body weight, white blood cell counts, blood hemoglobin, or platelet numbers after daily administration.14 Further analysis showed that following IP administration of 50, 75, or 100 mg/kg MP-A08 to mice for 14 days, no significant changes where observed in all immunophenotypically defined hematopoietic bone marrow lineages analyzed (supplemental Figure 5A).
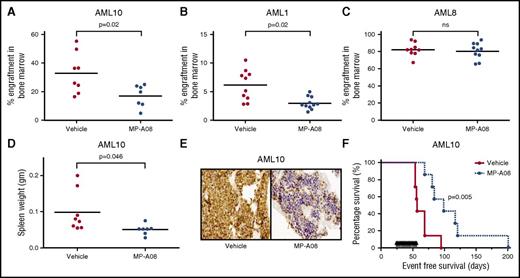
We next generated 3 separate sets of primary AML patient xenografts in NOD/SCID mice to test the efficacy of MP-A08 against this disease in vivo. Once significant disease was detected in the peripheral blood (≥5% human CD45+), administration of MP-A08 began at 100 mg/kg 6 times a week for 2 weeks. MP-A08 treatment significantly reduced leukemic burden in mice engrafted with AML1 and AML10. For AML8, there were no differences between the treated and untreated groups (Figure 4A-C). Analysis of the molecular mutations in the MP-A08–responsive xenografts (AML1 and AML10) revealed they both carry the FLT3-ITD mutation and either IDH1 or IDH2 (Table 1). The nonresponsive xenograft harbored the 11q23 MLL translocation, a cytogenetic risk group with poor overall survival.40 The xenografts retained CD33 expression demonstrating maintenance of their myeloid origin (supplemental Figure 5B), and splenomegaly was observed in the vehicle-treated mice compared with MP-A08–treated mice (Figure 4D). Immunohistochemistry on mouse bone marrow with mitochondrial antibodies (MTC02) specific for human cells confirmed that vehicle-treated mice displayed high levels of leukemic burden (brown cells) whereas MP-A08–treated mice had a reduced burden and a large proportion of mouse hematopoietic cells (purple cells) (Figure 4E).
In vivo therapeutic efficacy of MP-A08 in a human xenotransplantation model of AML. (A-C) NOD-SCID mice were engrafted with primary AML blasts, and after disease was established (2-6 weeks), mice were IP injected with either vehicle or MP-A08 (100 mg/kg) 6 times a week for 2 weeks. Engraftment was quantified by assessing the percentage of human CD45+ cells in the bone marrow of recipient mice. Each symbol represents the percentage of CD45+ cells observed in a separate mouse. Significance was assessed by Student t test. (D) Engrafted mice exhibited overt signs of AML including enlarged spleens. (E) Human cells in the bone marrow of engrafted mice were stained with the human specific mitochondrial antibody MTC02. (F) NOD-SCID mice were engrafted with primary AML blasts, and after disease was established, mice were injected IP 5 times/week for 32 days with 100 mg/kg MP-A08. P values were calculated using the log-rank Mantel-Cox test.
In vivo therapeutic efficacy of MP-A08 in a human xenotransplantation model of AML. (A-C) NOD-SCID mice were engrafted with primary AML blasts, and after disease was established (2-6 weeks), mice were IP injected with either vehicle or MP-A08 (100 mg/kg) 6 times a week for 2 weeks. Engraftment was quantified by assessing the percentage of human CD45+ cells in the bone marrow of recipient mice. Each symbol represents the percentage of CD45+ cells observed in a separate mouse. Significance was assessed by Student t test. (D) Engrafted mice exhibited overt signs of AML including enlarged spleens. (E) Human cells in the bone marrow of engrafted mice were stained with the human specific mitochondrial antibody MTC02. (F) NOD-SCID mice were engrafted with primary AML blasts, and after disease was established, mice were injected IP 5 times/week for 32 days with 100 mg/kg MP-A08. P values were calculated using the log-rank Mantel-Cox test.
To further determine if targeting SPHK1 conferred a survival advantage in AML, NOD/SCID mice with established disease following engraftment with primary AML patient cells where treated with MP-A08 at 100 mg/kg 5 days a week for 32 days. MP-A08 treatment significantly improved the event-free survival of mice with established leukemia compared with vehicle-treated control mice (median survival of 99 and 57 days, respectively; Figure 4F). Together these results demonstrate that targeting SPHK1 is effective against AML cells in vivo.
Targeting SPHK1 induces caspase-dependent cell death via inhibition of MCL1 and induction of BH3-only proteins
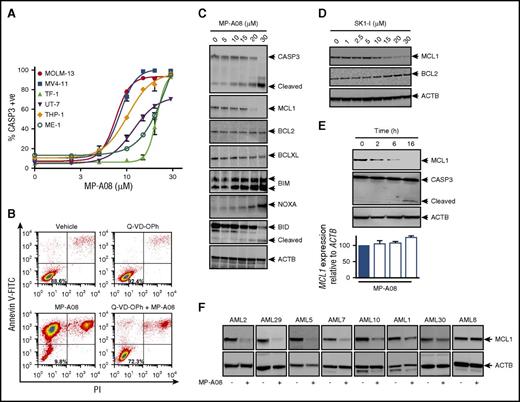
To characterize the cell death induced by MP-A08 in AML, we performed dose responses in a panel of AML cell lines and quantified caspase-3 activation, a hallmark of apoptosis. Caspase-3 activity increased in a dose-dependent manner across all cell lines examined (Figure 5A). We further characterized MV4-11 cells for early and late stages of apoptosis by annexin V and PI staining. MP-A08 treatment reduced cell viability (annexin V negative, PI negative) from 88.6% to 9.8% (Figure 5B) and induced caspase-3 cleavage (Figure 5C). Cell death occurred in a caspase-dependent manner with protection afforded by preincubation with the pan-caspase inhibitor Q-VD-OPh (Figure 5B).
SPHK1 inhibition induces caspase-dependent cell death through MCL1 suppression in AML cell lines and primary human AML blasts. (A) AML cell lines MOLM-13, MV4-11, TF-1, UT-7, THP-1, and ME-1 were treated with increasing doses of MP-A08 for 48 hours, and caspase-3 activity was measured by substrate cleavage of NucViewTM488 (Biotium) and quantified by flow cytometry. Results are mean ± range. (B) MV4-11 cells were preincubated for 1 hour with the pan-caspase inhibitor Q-VD-OPh (25 μM) and subsequently treated with MP-A08 (30 μM) for 24 hours prior to analysis of cell survival by annexin V/PI staining. MV4-11 cells were incubated with increasing doses of MP-A08 (C) or SK1-I (D) for 16 hours following which whole cell lysates were immunoblotted with the indicated antibodies. (E) MV4-11 cells were incubated with MP-A08 (20 μM) for the indicated time following which whole cell lysates were immunoblotted with the indicated antibodies or MCL1 mRNA levels determined using quantitative RT-PCR (mean ± SEM, n = 3). (F) Primary AML blasts were incubated with 30 μM of MP-A08 for 16 hours following which whole cell lysates were immunoblotted with the indicated antibodies.
SPHK1 inhibition induces caspase-dependent cell death through MCL1 suppression in AML cell lines and primary human AML blasts. (A) AML cell lines MOLM-13, MV4-11, TF-1, UT-7, THP-1, and ME-1 were treated with increasing doses of MP-A08 for 48 hours, and caspase-3 activity was measured by substrate cleavage of NucViewTM488 (Biotium) and quantified by flow cytometry. Results are mean ± range. (B) MV4-11 cells were preincubated for 1 hour with the pan-caspase inhibitor Q-VD-OPh (25 μM) and subsequently treated with MP-A08 (30 μM) for 24 hours prior to analysis of cell survival by annexin V/PI staining. MV4-11 cells were incubated with increasing doses of MP-A08 (C) or SK1-I (D) for 16 hours following which whole cell lysates were immunoblotted with the indicated antibodies. (E) MV4-11 cells were incubated with MP-A08 (20 μM) for the indicated time following which whole cell lysates were immunoblotted with the indicated antibodies or MCL1 mRNA levels determined using quantitative RT-PCR (mean ± SEM, n = 3). (F) Primary AML blasts were incubated with 30 μM of MP-A08 for 16 hours following which whole cell lysates were immunoblotted with the indicated antibodies.
The BCL2 family of proteins are well known to be critical determinants of cancer cell survival,41 with AML in particular having a high dependency on the prosurvival MCL1.42,43 Indeed, consistent with previous reports,44,45 we found MCL1 protein levels to be highly elevated in primary AML patient blasts, compared with bone marrow MNCs from healthy volunteers (supplemental Figure 6). To ascertain the mechanism by which SPHK1 inhibition induces mitochondrial-dependent apoptosis, we examined the changes in levels of MCL1 and other proapoptotic and prosurvival BCL2 family members after MP-A08 treatment. Importantly, we found a selective and significant reduction in the levels of the prosurvival MCL1 protein following SPHK1 inhibition with MP-A08 (Figure 5C). This was accompanied by increases in the proapoptotic BH3-only proteins BIM, NOXA, and cleaved BID (Figure 5C). Similar results were also observed following treatment of MV4-11 cells with the SPHK1-selective inhibitor SK1-I (Figure 5D), but not the SPHK2-selective inhibitor ABC294640 (supplemental Figure 7), confirming this inhibitory mechanism on MCL1 is mediated through SPHK1. Importantly, this loss of MCL1 occurred as early as 2 hours after SPHK1 inhibition, prior to caspase-3 activation, suggesting a contributory role to apoptosis, rather than a secondary effect (Figure 5E). Notably, quantitative RT-PCR analysis showed MCL1 mRNA levels remained unaltered following cell exposure to MP-A08 (Figure 5E), suggestive of posttranslational regulation of MCL1 protein levels. Further global RNA-Seq analysis of early gene expression changes, prior to caspase-3 activation, in response to MP-A08 also showed no change in MCL1 mRNA. A range of other genes did, however, show significant regulation (supplemental Figure 8A; supplemental Table 1), with pathway analysis suggesting SPHK1 inhibition in AML cells also altered the unfolded protein response and endoplasmic reticulum stress pathway, glucocorticoid receptor signaling, and cholesterol biosynthesis (supplemental Figure 8B), providing avenues for future exploration.
A survey of primary AML patient samples (n = 8) with heterogeneous mutational landscapes (Table 1) also demonstrated that MCL1 expression was lost upon MP-A08 treatment (Figure 5F). Notably, however, no loss of MCL1 was observed with MP-A08 in cells from patient AML8 (Figure 5F), consistent with the in vivo resistance of this AML to MP-A08 therapy (Figure 4C).
Targeting SPHK1 survival signaling synergizes with BH3 mimetics to induce cell death in AML
Because our findings suggested that SPHK1 inhibition induces apoptosis by targeting MCL1 expression in AML, we next examined if enforced MCL1 expression was sufficient to rescue MP-A08–induced cell death in these cells. Indeed, consistent with this hypothesis, doxycycline-inducible lentiviral expression of MCL1 in MV4-11 cells resulted in a partial, but significant increase in cell survival in the presence of MP-A08 (Figure 6A).
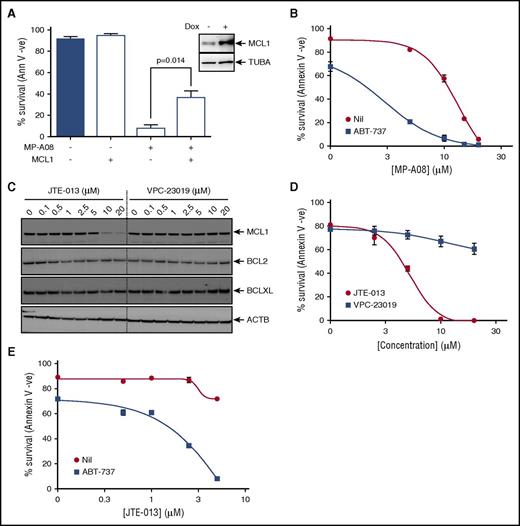
Targeting SPHK1 survival signaling synergizes with BH3 mimetics to induce cell death in AML. (A) MV4-11 cells were transduced with a doxycycline-inducible lentiviral vector to overexpress MCL1. Cells were then exposed to 1 µg/mL doxycycline and 24 hours postinduction treated with 30 μM of MP-A08 for 24 hours, and cell survival was determined by annexin V/PI staining. Results are mean ± SEM, n = 3. Immunoblot analysis of whole cell lysates with anti-MCL1 antibodies 24 hours postinduction confirmed doxycycline-inducible MCL1 expression. (B) MV4-11 cells were treated with increasing doses of MP-A08 in the presence (▪) or absence (●) of 10 nM ABT-737, and cell survival was quantified at 24 hours by annexin V/PI staining. Results are mean ± SEM, n = 3. (C) MV4-11 cells were treated with increasing doses of the S1PR2 antagonist JTE-013 or the S1PR1/3 antagonist VPC-23019 for 6 hours following which whole cell lysate were immunoblotted with the indicated antibodies. (D) MV4-11 cells were treated with increasing doses of JTE-013 (●) or VPC-23019 (▪), and cell survival was quantified at 48 hours by annexin V/PI staining. Results are mean ± SEM, n = 3. (E) MV4-11 cells were treated with increasing doses of JTE-013 in the presence (▪) or absence (●) of 10 nM ABT-737, and cell survival was quantified at 48 hours by annexin V/PI staining. Results are mean ± range. Significance was assessed by Student t test.
Targeting SPHK1 survival signaling synergizes with BH3 mimetics to induce cell death in AML. (A) MV4-11 cells were transduced with a doxycycline-inducible lentiviral vector to overexpress MCL1. Cells were then exposed to 1 µg/mL doxycycline and 24 hours postinduction treated with 30 μM of MP-A08 for 24 hours, and cell survival was determined by annexin V/PI staining. Results are mean ± SEM, n = 3. Immunoblot analysis of whole cell lysates with anti-MCL1 antibodies 24 hours postinduction confirmed doxycycline-inducible MCL1 expression. (B) MV4-11 cells were treated with increasing doses of MP-A08 in the presence (▪) or absence (●) of 10 nM ABT-737, and cell survival was quantified at 24 hours by annexin V/PI staining. Results are mean ± SEM, n = 3. (C) MV4-11 cells were treated with increasing doses of the S1PR2 antagonist JTE-013 or the S1PR1/3 antagonist VPC-23019 for 6 hours following which whole cell lysate were immunoblotted with the indicated antibodies. (D) MV4-11 cells were treated with increasing doses of JTE-013 (●) or VPC-23019 (▪), and cell survival was quantified at 48 hours by annexin V/PI staining. Results are mean ± SEM, n = 3. (E) MV4-11 cells were treated with increasing doses of JTE-013 in the presence (▪) or absence (●) of 10 nM ABT-737, and cell survival was quantified at 48 hours by annexin V/PI staining. Results are mean ± range. Significance was assessed by Student t test.
The BH3 mimetic ABT-737, which targets other prosurvival BCL2 family proteins, but not MCL1, has proved highly effective in tumors that have low dependency on MCL1 for cell survival.46-48 Some leukemias, including AML, however, appear resistant to ABT-737 because of elevated expression of MCL1,46 which is associated with poor patient survival.49 Therefore, we examined if targeting MCL1 levels via SPHK1 inhibition could sensitize AML cells to BH3 mimetics. Indeed, we found that subcytotoxic combinations of MP-A08 and ABT-737 induced synergistic cell death in AML cells (Figure 6B; CI <1).
Because many of the functions of SPHK1 are mediated via the engagement of S1P of its 5 cell surface receptors, of which we established AML cells largely express 3 (S1PR2-4; supplemental Figure 9A). Thus, we next used S1P receptor antagonists to determine the effects on the BCL2 prosurvival proteins in AML cells. Incubation of cells with VPC-23019, a selective antagonist of both S1PR1 and S1PR3,50 had no effect on the levels of BCL2 prosurvival proteins (Figure 6C). Antagonism of S1PR2 and S1PR4 with JTE-013,51,52 however, phenocopied the effects of SPHK1 inhibition by substantially reducing MCL1 expression without significant effects on BCL2 and BCLXL (Figure 6C). Notably, the S1PR4-selective antagonist CYM5035853 had no effect on MCL1 levels (supplemental Figure 9B) suggesting the observed effects of JTE-013 were mediated by antagonism of S1PR2. Consistent with its effects on MCL1, JTE-013 blocked cell survival in a dose-dependent manner in AML cells (Figure 6D), whereas neither VPC-23019 nor CYM50358 had effects on these cells (Figure 6D; supplemental Figure 9C). Finally, combination of ABT-737 with JTE-013 induced synergistic cell death in AML cells further supporting the notion that blocking SPHK1-mediated survival signaling via S1PR2 can sensitize AML cells to BH3 based mimetics (Figure 6E; CI <1).
Discussion
In this study, we assessed the role and targeting of SPHK1 in AML. Consistent with previous observations of SPHK1 overexpression in a diverse array of solid cancers,5 we found SPHK1 to be significantly overexpressed in AML. Importantly, the overexpression of SPHK1 was evident across a heterogeneous mutational landscape, suggesting that targeting SPHK1 may have broad utility across various AML subtypes. In support of this, total SPHK1 protein levels were elevated in all AML samples examined, irrespective of AML subtype or mutational landscape. Levels of phospho-SPHK1, the main oncogenic form of the enzyme,35 were also elevated in many AML biopsies. This is consistent with the common activation of ERK1/2, the kinases responsible for SPHK1 phosphorylation,34 in AML via diverse mutations in upstream ERK1/2-regulatory pathways, including RAS, receptor tyrosine kinases and cytokine receptor-associated kinases such as JAK1/2.54
We demonstrated that inhibition of SPHK1 with either MP-A08 or SKI-II induced cell death in an extended collection of AML patient blasts and isolated CD34+/CD38−/CD123+ LSPCs, with these inhibitors showing broad utility across various AML subtypes and mutational landscapes but little effect on normal CD34+ progenitors suggesting a potential therapeutic window for SPHK1 directed therapies.
Our studies with orthotopic xenografts of primary AML patient cells provides significant, physiologically relevant evidence that targeting SPHK1 with small molecules has anti-AML activity and considerably advances previous findings in this area with AML cell lines and nonhematological subcutaneous xenografts in mice.18-21 Indeed, MP-A08 treatment of mice engrafted with primary human AML cells significantly reduced leukemic burden and prolonged mouse survival without impacting on normal mouse hematopoiesis or blood, kidney, liver, heart, and spleen pathology.33 Notably, MP-A08 is known to be equally effective at inhibiting human and mouse sphingosine kinases.33 This therapeutic response occurred in 2 independent AML samples, both with normal karyotype, carrying the FLT3-ITD mutation and either IDH1 or IDH2 mutations, a common occurrence in AML,54 suggesting SPHK1-directed therapies may have utility across this spectrum of AML. Notably, cells from patient AML8, harboring the 11q23 MLL translocation, were nonresponsive to MP-A08 in vivo, consistent with our subsequent findings that SPHK1 inhibition in these cells did not result in loss of MCL1, a protein known to be critical for AML cell survival. This stability of MCL1 to SPHK1 inhibition did not, however, appear to be a consequence of the 11q23 MLL translocation, because all other AML patient cells examined that harbored this defect (AML2, AML7, and AML29) showed clear loss of MCL1 in response to SPHK1 inhibition (Figure 5F). With the known variability in this heterogeneous AML subtype,55 this suggests other unknown defects exist in AML8 that make these cells resistant to MP-A08–induced loss of MCL1.
The high mortality rates in AML are a consequence of disease relapse and subsequent chemotherapeutic resistance driven by a small reservoir of quiescent LSPCs that reside in niches within the bone marrow and are notoriously refractory to conventional chemotherapy regimens.36 In efforts to improve patient outcomes, new therapies have been developed whereby traditional genotoxic drugs have been combined with targeted therapies to block multiple cell survival signaling pathways and achieve synergistic cell death, and ultimately sustained disease remissions. A number of such therapies are currently in clinical trials for the treatment of AML, with promising results. For example, trials combining cytarabine with the polo-like kinase inhibitor volasertib,56 cyclin-dependent kinase 9 inhibitor flavopiridol,57,58 or the FLT3 inhibitors sorafenib59 or midostaurin60 have all produced promising clinical outcomes. Our findings that targeting SPHK1 with MP-A08 chemosensitized AML blasts and CD34+/CD38−/CD123+ LSPCs to cytarabine-induced synergistic cell death suggests that this may represent a promising approach for future combination therapies.
Considering MP-A08 induced caspase-dependent cell death in AML cells, we next surveyed the BCL2 family members to determine the mechanism of cell death. Our findings showing SPHK1 inhibitors induced loss of MCL1 in AML cells, whereas BCL2 and BCLXL were unaffected, are consistent with previous reports where in AML and chronic myeloid leukemia cell lines, targeting SPHK1 induced loss of MCL1 and reduced cell survival.19,61 Interestingly, however, MP-A08 treatment was also associated with induction of the proapoptotic BH3-only proteins BIM, NOXA, and cleaved BID, which was not observed previously when AML cell lines were exposed to another SPHK1 inhibitor, SKI-178.19 Notably, our findings demonstrate a role of S1PR2 in maintaining MCL1 expression because application of the S1PR2 antagonist JTE-013, but not antagonists of S1PR1, 3, and 4, induced a similar loss of MCL1 in AML cells to that observed with SPHK1 inhibitors. These results are consistent with the emerging oncogenic role of S1PR2 signaling in solid tumors and hematological malignancies62-64 providing independent validation of our findings with SPHK1 inhibitors and demonstrate that targeting the SPHK1/S1P axis has an important effect on regulating MCL1 expression and AML cell survival. Although the signaling pathways downstream of S1PR2 that mediate these effects are yet to be deciphered, in chronic myeloid leukemia activation of S1PR2 inhibits protein phosphatase 2A (PP2A), thereby blocking dephosphorylation of BCR-ABL and promoting stability of this oncogenic fusion protein.64 Because stability of MCL1 is also regulated by phosphorylation,65 it is tempting to speculate that such a mechanism may also operate to regulate MCL1 expression. Notably, however, inhibition of PP2A has been recently shown to enhance MCL1 degradation in lymphoma cells,66 suggesting the situation may be more complex.
The development of BH3 mimetics such as ABT-737 represents one of the most promising rationally designed drugs and the first example of drugging a protein-protein interaction.67 Despite the success of second-generation BH3 mimetics such as ABT-199 in the treatment of chronic lymphoblastic leukemia68 and AML,69 current BH3 mimetics fail to inhibit MCL1, which is fundamental to the pathogenesis of most forms of AML42,43 and the major mechanism of resistance to BH3 mimetic–based therapies.46,47 The development of a pan-BH3 mimetic termed obatoclax,70 which inhibited MCL1 in addition to BCL2 and BCLXL, induced neuronal toxicity.71,72 To circumvent the toxicities associated with targeting all prosurvival BCL2 family members with pan-BH3 mimetics, many groups have developed combinational targeted therapies whereby BCL2 selective inhibitors were administered in combination with inhibitors of MCL1. For example, sorafenib sensitized AML cells to ABT-73773 and obatoclax74 as does targeting phosphoinositide 3-kinase and mTOR in combination with BH3 mimetics.75 As we demonstrated that MCL1 overexpression rescued cell death induced by SPHK1 targeted therapies, and combinational therapies with ABT-737 and agents targeting the SPHK1/S1PR2 axis induced synergistic lethality in AML, these rational combinational therapies may prove efficacious for the treatment of AML. Targeting these 2 cooperative survival signaling pathways may have the capacity to overcome the diverse molecular landscapes associated with heterogeneous diseases such as AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are thankful to the patients and to the South Australian Cancer Research Biobank.
This work was supported by the Fay Fuller Foundation, the National Health and Medical Research Council of Australia, the Cancer Council of South Australia Beat Cancer Project, the Royal Adelaide Hospital Research Fund, and the Channel 7 Children’s Research Foundation.
Authorship
Contribution: J.A.P., A.C.L., W.Z., J.T., C.T.W.-B., P.A.B.M., H.S.R., N.C., and A.H.W. performed experiments; I.D.L. facilitated collection of AML biopsies; I.D.L., D.I., and S.E.S. collated clinical notes; M.R.P., D.T., A.F.L., R.J.D., and I.D.L. provided intellectual input and edited the manuscript; and J.A.P. and S.M.P. designed the studies, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stuart M. Pitson, Centre for Cancer Biology, SA Pathology Building, Frome Rd, Adelaide, SA 5000, Australia; e-mail: stuart.pitson@unisa.edu.au.

![Figure 2. Targeting SPHK1 induces AML cell death. AML cell lines (A), primary AML blasts (B), or normal bone marrow–derived CD34+ cells (C) were plated in the indicated concentrations of MP-A08, and cell survival was determined by annexin V staining. (D) FACS-purified primary human CD34+/CD38−/CD123+ LSPCs were treated with a dose response of MP-A08 for 24 hours, and cell survival was determined by annexin V staining. Results are mean ± range. MV4-11 cells were transduced with doxycycline-inducible short hairpin RNA (shRNA) constructs to knockdown SPHK1 and a nontargeting control shRNA. Cells were incubated with 1 μg/mL doxycycline (Dox) and 48 hours postinduction quantitative RT-PCR (mean ± standard deviation, n = 3) (E), SPHK1 activity assays (mean ± range) (F), immunoblot analysis of whole cell lysates were performed and quantified by laser densitometry (ratio of SPHK1/ACTB) (G), and analysis of cell survival was determined by annexin V staining (mean ± standard error of the mean [SEM], n = 3) (H). Significance was assessed by Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/6/10.1182_blood-2016-06-720433/4/m_blood720433f2.jpeg?Expires=1767728104&Signature=EUVkMHU8amh5aLziwG28XRCMB~tRehDFvzKD~OkQEaAFCKUn7PWG4PrAD0Rs8EGrXv-8AE1XTPUqxZlBEPeA8VFIWpTsXo5BTZ4G6uv0jvczi6p0rnBFF3o3Esp0AKw9peMpPhw9mvRsyQuYCnyvoA9Yfn9CUvRxgLjFj4yygaRopRiPN644FkiSEDX-XmLM1WYQw-oCB4ZPCHiQ6~mBsmbAvyFh8mwiqlt45dcgQMVmM64hcrOwuZ9u26MvGQNlZ0aXoL4fcmgiwgPoeLKirGj2m8JyfKyMir-6nRSj-08sj~wJIIIp0A~6L74E5QfC2UZAD3kiLVW2KanoRJtfPw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal