Key Points
ZEB2, a novel driver of immature T-ALL, interacts with the lysine-specific demethylase KDM1A.
KDM1A function is critical for leukemic survival of T-ALL cells with high ZEB2 levels.
Abstract
Elevated expression of the Zinc finger E-box binding homeobox transcription factor-2 (ZEB2) is correlated with poor prognosis and patient outcome in a variety of human cancer subtypes. Using a conditional gain-of-function mouse model, we recently demonstrated that ZEB2 is an oncogenic driver of immature T-cell acute lymphoblastic leukemia (T-ALL), a heterogenic subgroup of human leukemia characterized by a high incidence of remission failure or hematological relapse after conventional chemotherapy. Here, we identified the lysine-specific demethylase KDM1A as a novel interaction partner of ZEB2 and demonstrated that mouse and human T-ALLs with increased ZEB2 levels critically depend on KDM1A activity for survival. Therefore, targeting the ZEB2 protein complex through direct disruption of the ZEB2-KDM1A interaction or pharmacological inhibition of the KDM1A demethylase activity itself could serve as a novel therapeutic strategy for this aggressive subtype of human leukemia and possibly other ZEB2-driven malignancies.
Introduction
Zinc finger E-box binding homeobox transcription factor-2 (ZEB2) is a member of the Zinc finger E-box binding homeobox transcription factor family that mediates epithelial to mesenchymal transition (EMT) events during development and disease.1 Induced expression of ZEB2 in epithelial cancer cell lines results in the repression of a wide range of genes responsible for cellular adhesion, allowing these cells to become motile and upon xenotransplantation disseminate into the surrounding tissue and metastasize.2 Moreover, increased expression of EMT transcription factors (EMT-TFs), such as ZEB2, is associated with the acquisition of cancer stem cell (CSC) properties that have the potential to self-renew and form secondary tumors upon transplantation.3-5 Currently, little is known about how EMT-TFs regulate CSC properties at the molecular level. It has been proposed that targeting EMT-TFs is a promising novel therapeutic strategy that not only prevents EMT-mediated spreading of tumor cells but also targets radio-/chemoresistant CSCs.6
Using a conditional loss-of-function approach, we have demonstrated that ZEB2 is an essential transcription factor during embryonic and adult hematopoiesis.7,8 In contrast, conditional Zeb2 overexpression leads to the spontaneous formation of an immature early thymic progenitor subtype of T-cell acute lymphoblastic leukemia (ETP-ALL).5 ETP-ALL is a refractory and aggressive form of leukemia, characterized by the coexpression of early T-cell and myeloid progenitor cell gene expression profiles.9 Zeb2-overexpressing primary T-cell acute lymphoblastic leukemia (T-ALL) cells show significant overlap with the expression profile of human ETP-ALL, and exhibit a marked increase of hematopoietic stem cell (HSC) markers and leukemia-initiation potential.5
ZEB2 is a large multidomain homeobox transcription factor that recognizes bipartite E-box motifs through its amino- and carboxyterminal Zinc finger domains.10 The domains outside the Zn-finger clusters have been shown to be essential for the recruitment of various tissue-specific coactivators/repressors, which ultimately regulates ZEB2’s tissue-specific activity.11 Therefore, identification and targeting of novel interaction partners that are essential for ZEB2’s oncogenic properties in the context of T-ALL represents a feasible option for the development of novel therapeutics to treat aggressive leukemia.
Recent studies have shown the importance of epigenetic changes during cancer initiation/progression. Clonal evolution studies have suggested the existence of preleukemic epigenetic changes within hematopoietic progenitors that allows clonal expansion and accumulation of genetic lesions that eventually results in overt leukemia.12-14 KDM1A is a flavin-containing amino oxidase that specifically catalyzes the demethylation of mono- and dimethylated lysines on histone 3 (H3K4 and H3K9, typically associated with gene repression and activation, respectively). KDM1A regulates the balance between self-renewal and differentiation of pluripotent stem cells,15 and its expression is upregulated in various cancers. Pharmacological inhibition of KDM1A has emerged as a promising novel therapy to treat and kill CSCs and novel potent inhibitors are being tested in clinical trials.16,17 Within the hematopoietic system, conditional loss of KDM1A results in a pancytopenia with impaired HSC self-renewal and differentiation potential.18 Inversely, KDM1A gain of function results in enhanced self-renewal and skewing toward the T-cell lineage, eventually leading to the development of T-cell lymphoblastic leukemia.19 Although KDM1A inhibition has been identified as a promising novel epigenetic therapy for various subtypes of human cancers including acute myeloid leukemia (AML), the molecular mechanisms that drive susceptibility to KDM1A inhibition and/or biomarkers that could predict KDM1A sensitivity remain to be further explored.
Here, we identify KDM1A as a novel interaction partner for ZEB2 in T-ALL and demonstrate that increased ZEB2 expression can drive sensitivity toward KDM1A inhibition.
Methods
Pull-downs, mass spectrometry
Mouse T-ALL cells were washed once with phosphate-buffered saline, and nuclear extracts were prepared as described previously20 and in the supplemental Methods (available on the Blood Web site). Anti-FLAG M2 agarose beads (Sigma) were used to pull down FLAG-ZEB2 containing protein complexes overnight at 4°C, rotating. Beads were washed 5 times and eluted 4 times with 0.6 mg/mL FLAG tripeptide (Sigma) for 15 minutes at room temperature. Fractions were loaded onto a 10% Mini-Protean TGX precast gel (BioRad) and silver stained (Thermo Fisher Scientific). Elutions containing the majority of FLAG-ZEB2 were loaded onto a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel (Thermo Scientific) and separated by a short electrophoresis gel run. Entire lanes were excised and subjected to in-gel tryptic digestion and liquid chromatography tandem mass spectrometry analysis as previously described.21 Raw data were processed and searched by MaxQuant software (version 1.5.2.8).22 Database searching was done against mouse protein sequences downloaded from Uniprot (03/2015 release) using MaxQaunt default settings. Significance of specific protein–protein interactions was performed using Perseus analysis software.23
Coimmunoprecipitations
For FLAG-ZEB2 pull-downs, anti-FLAG M2 agarose beads (Sigma) were used as previously described. For KDM1A pull-downs, the rabbit polyclonal anti-KDM1A (LSD1) antibody (Abcam) was used in combination with the Rabbit TrueBlot IP-set (Rockland). Western blot analysis was performed according to standard protocols and primary antibodies are listed in the supplemental Methods.
ChIP
Chromatin immunoprecipitations (ChIPs) were performed using the SimpleChIP Enzymatic Chromatin IP kit with magnetic beads (Cell Signaling Technology) using H3K4me2-specific mouse monoclonal Antibody (MABI 0303, Active Motif). The enriched DNA was quantified by real-time polymerase chain reaction (PCR) with SimpleChIP Human CD11b Promoter Primers (#14271, Cell Signaling Technology).
Cell culture
T-ALL cell lines were grown as previously described5 and in the supplemental Methods. For the generation of KARPAS-45 cells overexpressing ZEB2, virus production was performed in HEK293TN cells using JetPEI polyplus with pMD2.G (envelope plasmid), psPAX2 (packaging plasmid), and pSIN-TRE-GW-3xHA hZEB2 (target plasmid) in 0.1/0.9/1 ratios. Transduced KARPAS-45 cells were selected by puromycin (1 µg/mL). Forty-eight hours before the start of the KDM1A inhibitor experiment, doxycyclin (1 µg/mL) administration was started. GSK2879552 (ActiveBiochem) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mg/mL and diluted further in medium as indicated. Further cell culture details are listed in supplemental Methods.
siRNA knockdown
For small interfering RNA (siRNA)–mediated knockdown experiments, LOUCY was electroporated with 400 nM of ZEB2-specific siRNAs (siZEB2-01: ON-TARGETplus Human ZEB2 siRNA J-006914-07, Dharmacon; siZEB2-02: Silencer Select Predesigned Human ZEB2 siRNA s19034, ThermoFisher Scientific) or 400 nM of scrambled siRNAs (ON-TARGETplus Nontargeting pool D-001810-10-05, Dharmacon). Also, electroporation without adding siRNAs was performed as control. For the electroporation, an exponential decay pulse (300 V, 1000 mF; Genepulser MxCell, Bio-Rad) was used. After 72 hours and 120 hours, RNA was isolated and evaluated by quantitative reverse transcription PCR.
Cell viability assays
Cell viability was determined by combination of CellTiter Glo Luminiscent viability assay (Promega) and flow cytometric analysis on LSRII or Fortessa (BD Biosciences) and FACSDiva or FlowJo software (BD Biosciences). For viability, we used combination of annexin V/4′,6-diamidino-2-phenylindole stain or the fixable viability Dye eFluro450 (eBioscience).
Quantitative reverse transcription PCR was performed according to standard protocols, and used primer sets are listed in the supplemental Methods.
Allo-/xenotransplantations
T-ALL cell lines were intravenously injected in NOD scid γ (NSG) mice. For the recipients transplanted with the mouse T-ALL cell lines, 1 × 106 cells were transplanted into 8- to 10-week-old male recipients via tail-vein injection, and the KDM1A inhibitor treatment was started the day after cell injection with 1.5 mg GSK2879552/kg body weight (or vehicle only) via oral gavage and was continued for 3 weeks. Mice were monitored for illness and leukemia development.
For the NSG mice injected with LOUCY or PEER cells, KDM1A inhibitor treatment was started when the first signs of engraftment were detected (hCD45+ cells in periphery). Mice were treated (1.5 mg GSK2879552/kg body weight or vehicle via oral gavage) for 17 (PEER) or 18 (LOUCY) consecutive days. Mice were weighed daily and monitored. The percentage of leukemic cells in the blood was analyzed by staining the cells with a phycoerythrin-labeled antibody (Miltenyi Biotec) for human CD45 (hCD45), red blood cell lysis and measuring the percentage with S3e cell sorter (Bio-Rad).
RNA sequencing (RNA-seq) and bioinformatics
RNA was prepared using either the RNeasy mini Plus kit (Qiagen) or the miRNeasy mini kit (Qiagen). Library preparation was performed using the Illumina TruSeq RNA sample Prep Kit. Enriched complementary DNA libraries were sequenced using an Illumina NextSequation 500 sequencer instrument. Reads were aligned to GRCm38 and GRCh38 using STAR2.4.2a. Genes were quantified on Ensembl38_v81 and Gencode v23, respectively. Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes using standard settings. Finally, preranked gene set enrichment analysis (GSEA) was performed with the c2 MSigDB gene sets collection using the GSEA desktop application (Broad Institute, version v2.2.0). Further details are provided in the supplemental Methods.
Results
ZEB2 interacts with KDM1A
We derived 4 T-ALL cell lines from murine thymomas from our Zeb2 overexpression mouse model that had been intercrossed onto a conditional p53-null tumor-prone background5 : 2 p53 null primary T-ALL cell lines with Zeb2 overexpression (further annotated as P53/R26-Zeb2tg/tg) and 2 p53 null control cell lines without Zeb2 overexpression (further annotated as P53/R26+/+). In the P53/R26-Zeb2tg/tg tumor cell lines, the ZEB2 transgene contains an aminoterminal 3xFLAG/Strep-tag, providing a useful in vitro model system to isolate ZEB2-containing protein complexes and identify ZEB2 interaction partners in the context of T-ALL. We performed protein complex pull-down experiments on nuclear extracts of P53/R26-Zeb2tg/tg and P53/R26+/+ T-ALL cell lines using anti-FLAG beads (Figure 1A), followed by mass spectrometry. Importantly, known ZEB2 interaction partners, such as CTBP124 and multiple subunits of the NuRD chromatin remodeling complex (ie, CHD4/Mi-2β, MTA2, and HDAC1),25 were copurified in the FLAG-ZEB2–expressing tumor lines, confirming the validity of our approach (Table 1). We also identified novel putative ZEB2 interaction partners including the hematopoietic transcription factor RUNX1, MYB binding protein 1A (MYBBP1A), multiple proteins involved in pre–messenger RNA (mRNA) processing and RNA biogenesis (Hnrnph1, Snrpb/n, Snrpd2, Snrnp70, Ddx21), NUMA1, and the lysine-specific demethylase-1 KDM1A (Table 1). Here, we focused on the ZEB2-KDM1A interaction for several reasons: (1) there is a significant overlap between the phenotypes observed after KDM1A and ZEB2 gain/loss in vitro (ie, EMT phenotype switching,10,26 regulation of the balance between self-renewal and differentiation in embryonic stem cells,15,27 and regulating proliferation and differentiation of AML cells28,29 ) and in vivo (ie, pancytopenia with loss of HSC differentiation potential upon loss of function7,18 and spontaneous T-ALL formation upon gain of function,5,19 association with EMT and solid tumor metastasis10,26 ); (2) KDM1A has previously been identified as being part of both the NuRD26 and CTBP1 corepressor complexes,30 known interactors of ZEB2 and also identified here; and (3) the existence of potent KDM1A inhibitors16,17,31 that are presently being used in phase 1 clinical trials.
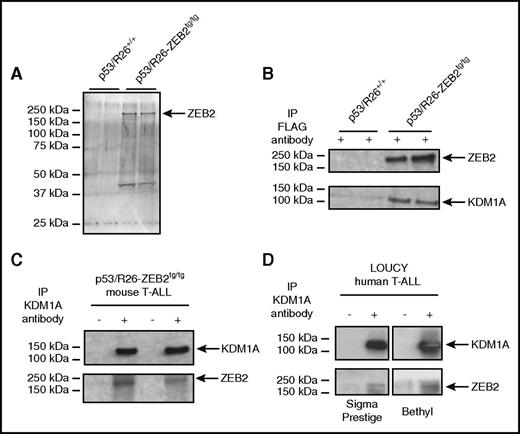
ZEB2 interacts with KDM1A in T-ALL cell lines. (A) Silver stain of SDS–polyacrylamide gel electrophoresis after pull downs of FLAG-ZEB2 protein complex. (B) Coimmunoprecipitation of KDM1A after pull down of FLAG-ZEB2 protein complex in mouse T-ALL cell lines. (C) Coimmunoprecipitation of ZEB2 after pull down of KDM1A protein complex in mouse T-ALL cell lines. (D) Coimmunoprecipitation of ZEB2 after pull down of KDM1A protein complex in the human T-ALL cell line LOUCY, using 2 antibodies that recognize human ZEB2.
ZEB2 interacts with KDM1A in T-ALL cell lines. (A) Silver stain of SDS–polyacrylamide gel electrophoresis after pull downs of FLAG-ZEB2 protein complex. (B) Coimmunoprecipitation of KDM1A after pull down of FLAG-ZEB2 protein complex in mouse T-ALL cell lines. (C) Coimmunoprecipitation of ZEB2 after pull down of KDM1A protein complex in mouse T-ALL cell lines. (D) Coimmunoprecipitation of ZEB2 after pull down of KDM1A protein complex in the human T-ALL cell line LOUCY, using 2 antibodies that recognize human ZEB2.
Mass spectrometric analysis of peptides identified after pull-down of FLAG-ZEB2 containing protein complexes in P53/R26-Zeb2tg/tg vs control P53/R26+/+ mouse T-ALL cell lines
| Protein name . | Gene name . | Peptides P53/R26+/+ control #1 . | Peptides P53/R26+/+ control #2 . | Peptides P53/Zeb2tg/tg TG/TG #1 . | Peptides P53/Zeb2tg/tg TG/TG #2 . |
|---|---|---|---|---|---|
| Gelsolin | Gsn | 0 | 0 | 21 | 6 |
| F-actin-capping protein subunit β | Capzb | 0 | 0 | 22 | 2 |
| Zinc finger E-box-binding homeobox 2 | Zeb2 | 0 | 0 | 13 | 4 |
| Major vault protein | Mvp | 0 | 0 | 10 | 3 |
| Nuclear mitotic apparatus protein 1 | Numa1 | 0 | 0 | 9 | 4 |
| Actin-related protein 2/3 complex subunit 4 | Arpc4 | 0 | 0 | 9 | 3 |
| Heterogeneous nuclear ribonucleoprotein H | Hnrnph1 | 0 | 0 | 8 | 3 |
| Small nuclear ribonucleoprotein-associated protein B/N | Snrpb;Snrpn | 0 | 0 | 9 | 1 |
| Histone deacetylase 1 | Hdac1 | 0 | 0 | 7 | 3 |
| Metastasis-associated protein MTA2 | Mta2 | 0 | 0 | 7 | 2 |
| Small nuclear ribonucleoprotein Sm D2 | Snrpd2 | 0 | 0 | 8 | 1 |
| Actin-related protein 3 | Actr3 | 0 | 0 | 8 | 1 |
| Myb-binding protein 1A | Mybbp1a | 0 | 0 | 5 | 2 |
| RuvB-like 1 | Ruvbl1 | 0 | 0 | 5 | 2 |
| Chromodomain-helicase-DNA-binding protein 4/5 | Chd4;Chd5 | 0 | 0 | 6 | 1 |
| U1 small nuclear ribonucleoprotein 70 kDa | Snrnp70 | 0 | 0 | 5 | 1 |
| Runt-related transcription factor 1 | Runx1 | 0 | 0 | 5 | 1 |
| Tropomyosin α-3 chain | Tpm3 | 0 | 0 | 4 | 2 |
| Lysine-specific histone demethylase 1A | Kdm1a | 0 | 0 | 4 | 1 |
| C-terminal-binding protein 1 | Ctbp1 | 0 | 0 | 3 | 3 |
| Spectrin β chain, nonerythrocytic 1 | Sptbn1 | 0 | 0 | 4 | 2 |
| Nucleolar RNA helicase 2 | Ddx21 | 0 | 0 | 3 | 2 |
| Unconventional myosin-Ic | Myo1c | 0 | 0 | 4 | 1 |
| Protein name . | Gene name . | Peptides P53/R26+/+ control #1 . | Peptides P53/R26+/+ control #2 . | Peptides P53/Zeb2tg/tg TG/TG #1 . | Peptides P53/Zeb2tg/tg TG/TG #2 . |
|---|---|---|---|---|---|
| Gelsolin | Gsn | 0 | 0 | 21 | 6 |
| F-actin-capping protein subunit β | Capzb | 0 | 0 | 22 | 2 |
| Zinc finger E-box-binding homeobox 2 | Zeb2 | 0 | 0 | 13 | 4 |
| Major vault protein | Mvp | 0 | 0 | 10 | 3 |
| Nuclear mitotic apparatus protein 1 | Numa1 | 0 | 0 | 9 | 4 |
| Actin-related protein 2/3 complex subunit 4 | Arpc4 | 0 | 0 | 9 | 3 |
| Heterogeneous nuclear ribonucleoprotein H | Hnrnph1 | 0 | 0 | 8 | 3 |
| Small nuclear ribonucleoprotein-associated protein B/N | Snrpb;Snrpn | 0 | 0 | 9 | 1 |
| Histone deacetylase 1 | Hdac1 | 0 | 0 | 7 | 3 |
| Metastasis-associated protein MTA2 | Mta2 | 0 | 0 | 7 | 2 |
| Small nuclear ribonucleoprotein Sm D2 | Snrpd2 | 0 | 0 | 8 | 1 |
| Actin-related protein 3 | Actr3 | 0 | 0 | 8 | 1 |
| Myb-binding protein 1A | Mybbp1a | 0 | 0 | 5 | 2 |
| RuvB-like 1 | Ruvbl1 | 0 | 0 | 5 | 2 |
| Chromodomain-helicase-DNA-binding protein 4/5 | Chd4;Chd5 | 0 | 0 | 6 | 1 |
| U1 small nuclear ribonucleoprotein 70 kDa | Snrnp70 | 0 | 0 | 5 | 1 |
| Runt-related transcription factor 1 | Runx1 | 0 | 0 | 5 | 1 |
| Tropomyosin α-3 chain | Tpm3 | 0 | 0 | 4 | 2 |
| Lysine-specific histone demethylase 1A | Kdm1a | 0 | 0 | 4 | 1 |
| C-terminal-binding protein 1 | Ctbp1 | 0 | 0 | 3 | 3 |
| Spectrin β chain, nonerythrocytic 1 | Sptbn1 | 0 | 0 | 4 | 2 |
| Nucleolar RNA helicase 2 | Ddx21 | 0 | 0 | 3 | 2 |
| Unconventional myosin-Ic | Myo1c | 0 | 0 | 4 | 1 |
We confirmed this ZEB2-KDM1A interaction using bidirectional coimmunoprecipitation experiments, with FLAG/ZEB2- and KDM1A-specific antibodies (Figure 1B-C). Finally, to prove that this interaction was not an overexpression artifact in our mouse T-ALL cell lines, we further confirmed this interaction in nuclear extracts from LOUCY cells (Figure 1D), a human T-ALL cell line with an immature expression profile and high ZEB2 expression.5
Mouse T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibitors
To validate the functional relevance of this ZEB2-KDM1A interaction, we tested the effects of KDM1A inhibition on the mouse T-ALL cell lines with and without Zeb2 overexpression. Strikingly, both Zeb2-overexpressing P53/R26-Zeb2tg/tg cell lines showed high sensitivity toward KDM1A inhibition (Figure 2A; supplemental Figure 1). More specifically, T-ALL cells with enhanced Zeb2 levels underwent apoptosis 48 hours after the start of the treatment with an average 50% inhibitory concentration at day 4 of 10.39 nM for the commercially available KDM1A inhibitor GSK2879552 (Figure 2A; supplemental Figure 1). In contrast, no effects were observed for the control P53/R26+/+ mouse T-ALL cell lines using these inhibitors (Figure 2A; supplemental Figure 1). Both P53/R26+/+ and P53/R26-Zeb2tg/tg expressed similar protein levels of KDM1A, illustrating that altered levels are not determining their sensitivity toward pharmacological KDM1A inhibition (supplemental Figure 2A).
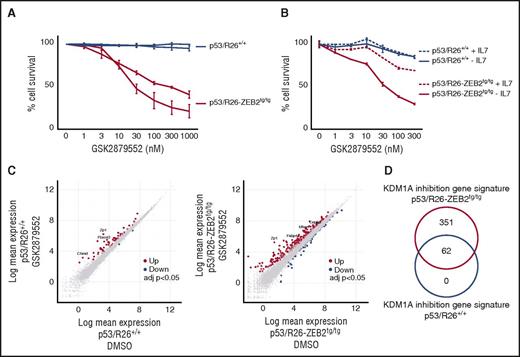
Mouse T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibition. (A) Leukemic cell survival of P53/R26-Zeb2tg/tg Zeb2 overexpressing mouse T-ALL cell lines compared with control P53/R26+/+ cell lines treated with different concentrations of the KDM1A inhibitor GSK2879552 for 4 days. The average of 3 independent experiments and standard deviation is plotted. (B) Effects of exogenous IL-7 on the KDM1A inhibitor-induced cell death observed in P53/R26-Zeb2tg/tg Zeb2 overexpressing mouse T-ALL cell lines. Average of technical triplicates are plotted. Similar effects are observed with the second independent mouse T-ALL cell lines. (C) Differential gene expression after KDM1A inhibition (24 hours, 100 nM GSK2879552) in P53/R26+/+ and P53/R26-Zeb2tg/tg mouse T-ALL cell lines (adjusted P value <.05, triplicates). (D) Overlap between the KDM1A responsive gene signatures in P53/R26+/+ and P53/R26-Zeb2tg/tg mouse T-ALL cell lines.
Mouse T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibition. (A) Leukemic cell survival of P53/R26-Zeb2tg/tg Zeb2 overexpressing mouse T-ALL cell lines compared with control P53/R26+/+ cell lines treated with different concentrations of the KDM1A inhibitor GSK2879552 for 4 days. The average of 3 independent experiments and standard deviation is plotted. (B) Effects of exogenous IL-7 on the KDM1A inhibitor-induced cell death observed in P53/R26-Zeb2tg/tg Zeb2 overexpressing mouse T-ALL cell lines. Average of technical triplicates are plotted. Similar effects are observed with the second independent mouse T-ALL cell lines. (C) Differential gene expression after KDM1A inhibition (24 hours, 100 nM GSK2879552) in P53/R26+/+ and P53/R26-Zeb2tg/tg mouse T-ALL cell lines (adjusted P value <.05, triplicates). (D) Overlap between the KDM1A responsive gene signatures in P53/R26+/+ and P53/R26-Zeb2tg/tg mouse T-ALL cell lines.
To obtain molecular insights into the pathways involved in ZEB2-driven KDM1A inhibitor sensitivity, we subsequently performed RNA-seq on 2 P53/R26+/+ and 2 P53/R26-Zeb2tg/tg cell lines. As previously described,5 this analysis revealed that the P53/R26+/+ and P53/R26-Zeb2tg/tg tumor lines displayed a very distinct gene expression signature under baseline (DMSO) conditions, with 6675 significantly differentially expressed genes (adjusted P value <.05; supplemental Figure 3; supplemental Table 1). Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed increased expression of AML related transcripts, decreased expression of NOTCH pathway genes involved in T-cell differentiation combined with activation of JAK-STAT/MAPK signaling in ZEB2-derived tumor lines (supplemental Table 2). These differential gene patterns are in line with our previous observations that Zeb2 driven thymomas reflect an immature T-ALL subtype with increased stem cell markers and activated interleukin-7 receptor (IL-7R)/JAK-STAT signaling.5 Given the prosurvival effects of activated IL-7R/JAK-STAT signaling in the context of Zeb2-driven T-cell leukemogenesis, we wondered whether exogenous IL-7 could partially rescue the apoptotic phenotype observed in these cells. Indeed, analysis of Zeb2 transgenic cells in IL-7–supplemented culture conditions revealed that IL-7 inclusion could partially block KDM1A inhibitor-induced cell death in the P53/R26-Zeb2tg/tg cell lines (Figure 2B).
Next, we analyzed the transcriptional response upon short-term KDM1A inhibition (100 nM GSK2879552 for 24 hours) in the absence of exogenous IL-7 in both murine tumor entities. Notably, and in line with the drug response phenotypes, KDM1A inhibition in control P53/R26+/+ cell lines only caused limited effects on overall transcription with 62 differentially expressed genes (adjusted P value <.05; Figure 2C). In contrast, gene transcription was more severely affected by KDM1A perturbations in the P53/R26-Zeb2tg/tg lines with significant changes in the expression of 413 transcripts (315 up- and 98 downregulated; adjusted P value <.05; Figure 2C; supplemental Table 3). Notably, all 62 genes that showed differential expression upon KDM1A inhibition in P53/R26+/+ cell lines, were also altered in the P53/R26-Zeb2tg/tg tumor lines (Figure 2D). All together, these results demonstrate that Zeb2 overexpression modulates murine T-ALL responsiveness to KDM1A inhibitors.
Human T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibitors
To study the human relevance of our findings, we subsequently evaluated the putative correlation between ZEB2 expression levels and KDM1A inhibitor sensitivity in a panel of human T-ALL cell lines. Interestingly, human T-ALL cell lines characterized by high (LOUCY) or moderate (PEER) ZEB2 expression (Figure 3A) were sensitive toward KDM1A inhibition with average 50% inhibitory concentration values at day 12 of 112.91 nM (LOUCY) and 43.83 nM (PEER) for GSK2879552 (Figure 3B). In addition, a time-course experiment in LOUCY cells revealed that the first overt effects on leukemic cell survival appear at ∼5 to 7 days of KDM1A inhibition treatment (supplemental Figure 4). Moreover, this KDM1A inhibitor sensitivity was found to be similar to what has been described for AML cell lines31 (supplemental Figure 5). In contrast, the survival or proliferation of ZEB2-negative T-ALL cell lines, such as JURKAT, TALL-1, and ALL-SIL, was not affected by KDM1A inhibition (Figure 3B). Nevertheless, and in line with recent work,31 screening of additional T-ALL cell lines revealed that 2 other ZEB2-negative T-ALL cell lines, RPMI-8402 and HSB-2, were also highly sensitive to KDM1A inhibition, suggesting that alternative molecular mechanisms, independent of ZEB2, can also drive sensitivity toward KDM1A inhibition in the context of human T-ALL. Similarly, as seen in the mouse cell lines, no correlation was observed in the human T-ALL cell lines between KDM1A protein expression and KDM1A inhibitor sensitivity (supplemental Figure 2B).
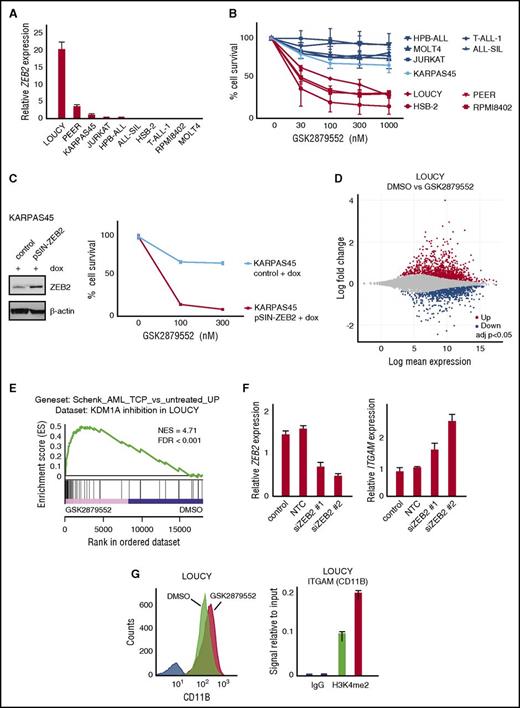
Human T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibition. (A) Relative mRNA expression in a panel of human T-ALL cell lines analyzed by quantitative PCR. (B) Cell viability of human T-ALL cell lines with variable expression of ZEB2 mRNA after 12 days administration of the KDM1A inhibitor GSK2879552. The average and standard deviation of 2 independent experiments is plotted. (C) Cell viability of KARPAS-45 with doxycycline (dox)–inducible overexpression of ZEB2 vs parental line after 12 days of GSK2879552 administration. Western blot demonstrating level of dox-inducible ZEB2 overexpression (left); % of cell viability (right). (D) Differential gene expression after KDM1A inhibition (48 hours, 100 nM GSK2879552) in LOUCY (adjusted P value <.05, triplicates). (E) Preranked GSEA using probe set of genes upregulated upon KDM1A inhibition by tranylcypromine (TCP) in human AML cell lines.32 (F) ZEB2 and ITGAM mRNA expression levels upon siRNA-mediated ZEB2 knockdown in LOUCY cells (100 nM GSK2879552, 72 hours). (G) Analysis of CD11B protein expression by flow cytometry after KDM1A inhibition (100 nM GSK2879552, 72 hours) correlated with increased H3K4me2 levels at ITGAM promoter analyzed by ChIP.
Human T-ALL cell lines with high ZEB2 levels are sensitive to KDM1A inhibition. (A) Relative mRNA expression in a panel of human T-ALL cell lines analyzed by quantitative PCR. (B) Cell viability of human T-ALL cell lines with variable expression of ZEB2 mRNA after 12 days administration of the KDM1A inhibitor GSK2879552. The average and standard deviation of 2 independent experiments is plotted. (C) Cell viability of KARPAS-45 with doxycycline (dox)–inducible overexpression of ZEB2 vs parental line after 12 days of GSK2879552 administration. Western blot demonstrating level of dox-inducible ZEB2 overexpression (left); % of cell viability (right). (D) Differential gene expression after KDM1A inhibition (48 hours, 100 nM GSK2879552) in LOUCY (adjusted P value <.05, triplicates). (E) Preranked GSEA using probe set of genes upregulated upon KDM1A inhibition by tranylcypromine (TCP) in human AML cell lines.32 (F) ZEB2 and ITGAM mRNA expression levels upon siRNA-mediated ZEB2 knockdown in LOUCY cells (100 nM GSK2879552, 72 hours). (G) Analysis of CD11B protein expression by flow cytometry after KDM1A inhibition (100 nM GSK2879552, 72 hours) correlated with increased H3K4me2 levels at ITGAM promoter analyzed by ChIP.
Although previous studies have suggested that KDM1A prevents differentiation and apoptosis in the context of MLL rearranged leukemia,28 we only observed minimal antileukemic effects upon GSK2879552 treatment in the MLL-AFX positive T-ALL cell line KARPAS-45 (Figure 3B). Given that this MLL rearranged T-ALL cell line is characterized by low levels of ZEB2 (Figure 3A), we subsequently generated KARPAS45 cells with doxycycline-inducible expression of ZEB2, to formally prove that ZEB2 can truly modulate KDM1A inhibitor sensitivity in the context of human T-ALL. Notably, doxycycline-induced ZEB2 overexpression significantly increased KDM1A inhibitor sensitivity in this in vitro model system, as exemplified by decreased cell viability 12 days after the initiation of KDM1A inhibition (Figure 3C).
Next, we analyzed the transcriptional response of KDM1A inhibition in human T-ALL cell lines. For this, RNA-seq was performed on biological triplicates of the ZEB2-positive LOUCY and PEER cells, and the ZEB2-negatives RPMI-8402 and HSB-2 treated for 48 hours with 100 nM GSK2879552 vs their corresponding DMSO treatment controls. Gene expression was affected by KDM1A inhibition in all sensitive T-ALL cell lines, but the effect was most pronounced in ZEB2-high LOUCY cells (supplemental Figure 6A). Indeed, GSK2879552 treatment in LOUCY cells resulted in significant changes of 2800 transcripts (1382 up- and 1418 downregulated; adjusted P value <.05, Figure 3D), whereas PEER, RPMI8402, and HSB-2 only displayed significant transcriptional changes in 333, 769, and 622 genes, respectively (supplemental Table 4). In terms of overlap, only 41 genes were commonly differentially regulated in all 4 T-ALL cell lines (39 genes up and 2 genes down; supplemental Figure 6B). Of note, cross-species comparison between LOUCY and P53/R26-Zeb2tg/tg murine tumor lines revealed a common KDM1A inhibitor response signature, which consisted of 87 genes (72 up and 15 down; supplemental Figure 7).
Interestingly, preranked GSEA revealed a significant overlap between the transcriptional response after KDM1A inhibition in the human LOUCY T-ALL cell line and the transcriptional response previously reported in the context of human AML32 (Figure 3E). For example, common responsive genes that are reexpressed upon KDM1A inhibition in both AML and LOUCY included GFI1, VIM, and ITGAM (encoding CD11B). Interestingly, recent work also demonstrated that loss of ZEB2 causes a similar reinduction of ITGAM expression in both mouse and human AML cell lines.29 Most notably, siRNA-mediated ZEB2 knockdown experiments in the T-ALL cell line LOUCY also resulted in increased ITGAM expression (Figure 3F). In line with this notion, we showed that CD11b induction upon KDM1A inhibition in LOUCY cells is associated with increased binding of the H3K4me2 mark at the ITGAM promoter region, as evaluated by ChIP (Figure 3G).
In vivo activity of KDM1A inhibition on high Zeb2 expressing murine and human T-ALL
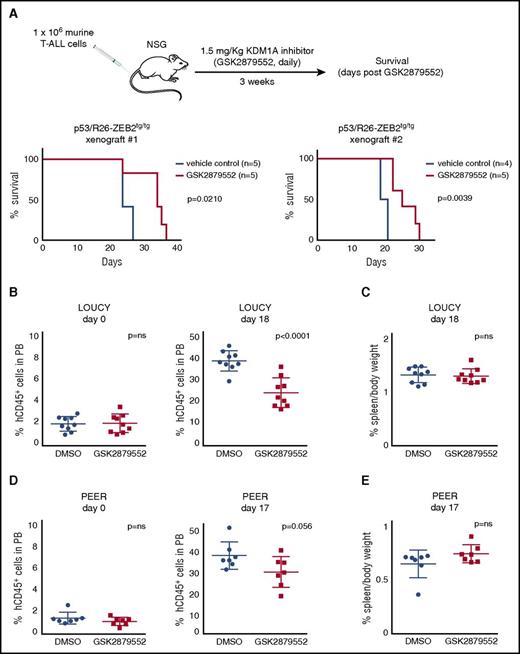
Given that promising in vivo responses have been reported for small cell lung cancer using the commercially available KDM1A inhibitor GSK2879552,17 we subsequently explored the in vivo potential of GSK2879552 to prevent or delay secondary leukemia formation of ZEB2-overexpressing murine T-ALL cell lines. For this, Zeb2-overexpressing T-ALL cell lines were xenografted in immune deficient NSG mice and treated for 3 weeks with GSK2879552 (1.5 mg/kg bodyweight) or with vehicle (DMSO). Notably, although all mice eventually developed leukemia, KDM1A inhibition triggered a significant delay in tumor latency (Figure 4A).
In vivo activity of KDM1A inhibitors on high Zeb2 expressing murine and human T-ALL. (A) Kaplan-Meier survival curves of NSG mice injected with 2 P53/R26-Zeb2tg/tg murine tumor lines with and without 3 weeks of GSK2879552 administration (1.5 mg/kg bodyweight). (B) Percentage of hCD45+ T-ALL cells in peripheral blood of NSG mice xenotransplanted with LOUCY human T-ALL cells, before and after 18 days of GSK2879552 administration (1.5 mg/kg bodyweight) vs DMSO. (C) Percent spleen weight after 18 days of KDM1A inhibition (1.5 mg GSK2879552/kg bodyweight) vs DMSO. (D) Percentage of hCD45+ T-ALL cells in peripheral blood of NSG mice xenotransplanted with PEER human T-ALL cells, before and after 17 days of GSK2879552 administration (1.5 mg/kg bodyweight) vs DMSO. (E) Percent spleen weight after 17 days of KDM1A inhibitor administration (1.5 mg/kg bodyweight) vs DMSO.
In vivo activity of KDM1A inhibitors on high Zeb2 expressing murine and human T-ALL. (A) Kaplan-Meier survival curves of NSG mice injected with 2 P53/R26-Zeb2tg/tg murine tumor lines with and without 3 weeks of GSK2879552 administration (1.5 mg/kg bodyweight). (B) Percentage of hCD45+ T-ALL cells in peripheral blood of NSG mice xenotransplanted with LOUCY human T-ALL cells, before and after 18 days of GSK2879552 administration (1.5 mg/kg bodyweight) vs DMSO. (C) Percent spleen weight after 18 days of KDM1A inhibition (1.5 mg GSK2879552/kg bodyweight) vs DMSO. (D) Percentage of hCD45+ T-ALL cells in peripheral blood of NSG mice xenotransplanted with PEER human T-ALL cells, before and after 17 days of GSK2879552 administration (1.5 mg/kg bodyweight) vs DMSO. (E) Percent spleen weight after 17 days of KDM1A inhibitor administration (1.5 mg/kg bodyweight) vs DMSO.
To further evaluate the therapeutic relevance of KDM1A inhibition in the context of human T-ALL, we subsequently performed in vivo drug treatment experiments using T-ALL cell line xenografts. LOUCY (ZEB2 high) and PEER (ZEB2 moderate) were injected in the tail vein of NSG mice, and treatment of established tumors (1.5 mg GSK2879552/kg bodyweight or vehicle) was initiated when the percentage of hCD45+ cells in peripheral blood reached 2% (3 weeks [PEER] and 4 weeks [LOUCY] after injection). Notably, KDM1A inhibition resulted in a decrease in the percentage of circulating hCD45+ for both cell lines, suggesting that GSK2879552 can trigger an in vivo effect on tumor burden and leukemia development (Figure 4B,D). However, no changes in spleen weight (Figure 4C,E) were observed and all animals showed signs of progressive disease, suggesting that KDM1A inhibition in human T-ALL might be particularly useful as part of a combination therapy.
Discussion
Relapsed T-ALL is a major clinical challenge. Therefore, current research efforts are focused on the development of more effective and less toxic antileukemic drugs, which will likely require an improved understanding of the molecular biology of chemotherapy-resistant residual tumor cells that eventually drive disease recurrence.33,34 In particular, immature T-ALL patients, which have a unique gene expression signature enriched for transcripts found in HSCs,35 have been associated with a poor response to current chemotherapy and significantly higher incidence of induction failure and hematological relapse.9
We recently demonstrated that ZEB2 is upregulated in a subset of human T-ALLs and that ZEB2 activation in the mouse results in the spontaneous formation of immature T-cell lymphoblastic leukemia with increased CSC properties and enhanced IL-7R expression.5 Therefore, determining ZEB2 interaction partners involved in this oncogenic driver role may open up novel therapeutic strategies to specifically target aggressive forms of leukemia.
Here, we identified a number of novel putative ZEB2 interaction partners, including recurrently mutated proteins in T-ALL, such as the essential hematopoietic transcription factor RUNX1,35 the RNA processing protein HNRNPH1,36 and NUMA1, a protein involved in regulation of mitotic spindle organization and asymmetric/symmetric cell divisions.37 More research is needed to confirm these novel ZEB2 interactions and to determine whether they are involved in ZEB2-mediated T-ALL formation. The identification of known ZEB2 interaction partners in this study, such as CTBP138 and the NuRD chromatin remodeling complex,25,39 confirms the validity of this approach and highlights their potential importance in T-ALL.
In this report, we identified and confirmed the interaction of ZEB2 with the lysine-specific histone demethylase KDM1A in T-ALL cells. Whether this interaction is direct or part of a larger complex remains to be determined. Nevertheless, given that KDM1A has no DNA-binding capacity, it depends on the interaction with transcription factors and transcriptional complexes to perform its function as a locus-specific epigenetic modifier. In that context, KDM1A has previously been identified in the CTBP130 and NuRD corepressor26 complexes, suggesting that these transcriptional complexes could also be functionally relevant in the context of T-cell transformation.
Similar as for ZEB2, KDM1A has been found to be involved in regulating cellular differentiation/self-renewal properties and EMT.26,40 Also other EMT-TF have been shown to interact with KDM1A and recruit it as a molecular hook to their target promoters,41 suggesting the presence of common regulatory mechanisms. Targeting either these interactions or the activity of KDM1A itself may therefore provide therapeutic avenues to prevent EMT and/or target chemo-/radioresistant CSCs.
Here, we demonstrated that mouse and human T-ALL cell lines with high Zeb2 mRNA expression levels are sensitive to KDM1A inhibition. Exactly how ZEB2 is capable of modulating KDM1A activity in the context of T-ALL remains to be determined. In AML cell lines, it was observed that KDM1A inhibition results in terminal differentiation by reactivation of the all-trans-retinoic acid pathway, leading to a severe decline in proliferative capacity. Indeed, Li et al29 recently demonstrated that high ZEB2 levels are essential for AML progression and loss of ZEB2 in mouse and human AML cells results in myeloid differentiation, similar to what has been observed for KDM1A inhibition. Also in AML cells, a ZEB2-KDM1A interaction could be demonstrated via coimmunoprecipitation experiments, thereby confirming this interaction in a separate leukemic setting. Interestingly, we also observed that KDM1A inhibition results in upregulation of the myeloid differentiation marker CD11B in the human T-ALL cell line LOUCY, suggesting overlapping molecular mechanism downstream of KDM1A inhibition in both leukemic settings. Nevertheless, our study also revealed that ZEB2-negative human T-ALL cell lines also showed strong KDM1A inhibitor sensitivity, suggesting that other mechanisms or oncogenic drivers/transcriptional factors, such as GFI1, might be able to bind and alter KDM1A activity and drive KDM1A inhibitor sensitivity in a ZEB2-independent manner.41,42
In this study, we have used a commercially available KDM1A inhibitor, GSK2879552, which was previously successfully used to analyze the in vivo effects of KDM1A inhibition on human small cell lung cancer xenografts. Here, in the context of T-ALL, we only observed a partial reduction of the leukemic burden in vivo. Therefore, T-ALL cell lines might need higher dosing or combinations with other chemotherapeutic agents (eg, all-trans-retinoic acid or HDAC inhibitors), as reported for other tumor entities.43-45 The differences observed between the therapeutic efficacy of KDM1A inhibition in vitro and in vivo, suggests that paracrine factors might regulate compensatory pathways that prevent cell death upon KDM1A inhibition in vivo. Here, we demonstrate that exogenous IL-7 can partially block KDM1A inhibitor-induced cell death in Zeb2 transgenic T-ALL cells. In line with this finding, human LOUCY T-ALL cells showed a similar up regulation of IL-7R expression following KDM1A inhibition. Therefore, combination therapies inhibiting both KDM1A and the IL-7R/JAK-STAT signaling pathway, for example, using the JAK inhibitor ruxolitinib, or inhibitors of the downstream effector BCL2, like ABT-199, may in vivo be more effective as compared with KDM1A monotherapy.
It remains to be determined whether or not the KDM1A interaction is also important for the phenotypes observed upon ZEB2 loss in adult HSCs. These include multilineage differentiation blocks with skewing to the granulocyte lineage and a myeloproliferative disorder–like phenotype with perturbed cytokine receptor signaling pathways.8 Similarly, it is not yet known whether this interaction is essential for other recently demonstrated roles of ZEB2 in cytotoxic T-cell differentiation,46,47 natural killer terminal maturation,48 and dendritic cell differentiation,49 as well as in other nonhematopoietic cell lineages such as melanocytes,50 neurons, and oligodendroglial cell fate and migration where Zeb2 has been previously demonstrated to play important roles.51-53
To conclude, we have identified a novel ZEB2-KDM1A interaction in the context of T-ALL, which is essential for the survival of ZEB2-overexpressing cells. Also in human T-ALL cell lines we see increased KDM1A inhibitor sensitivity upon enforced ZEB2 expression, suggesting a putative novel therapeutic tool for the treatment of ZEB2-driven tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Raymond Poot and Danny Huylebroeck for their advice on protein complex pull down experiment and Trevor Wilson for his help with RNA-seq. The authors also thank Béatrice Lintermans and Lindy Reunes for excellent technical assistance.
This work was supported by the Fund for Scientific Research– Flanders (FWO-V) (grants G056813N [J.J.H.]; KaN 1500416N [S.G.]; GA00113N, 3G065614, G0C4713N, and 31500615W [P.V.V.]; G052912N and G081713N [G.B.]), the Belgian Federation for the Study Against Cancer (grants 365W3415W [P.V.V.], B/13590 [G.B.], and 2016-121 [S.G.]), Belgian Stand Up to Cancer Foundation (grant 365Y9115W [P.V.V.]), Worldwide Cancer Research (grant 16-1157 [P.V.V., S.G., and J.J.H.]), the Swiss Bridge Award (P.V.V. and S.G.), and the National Health and Medical Research Council of Australia (grants 1047995 and 1104441 [J.J.H.]) and was part of the DevRepair (P7/07) IAP-VII network (S.G., J.J.H.). S.G. is postdoctoral fellow, and S.P. is a student supported by the FWO-V. N.V. and S.P. are students funded by the Belgian Stand Up to Cancer Foundation (grant 365Z1215W and Emmanuel van der Schueren grant). The computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by Ghent University, the Hercules Foundation, and the Department of Economy, Science and Innovation of the Flemish Government.
Authorship
Contribution: S.G., S.P., P.V.V., and J.J.H. designed the studies; S.G., S.P., K.H., T.N., S.E.S., M.C., C.C., J.W., M.T., and F.M. performed the experiments; S.G., S.P., W.V.L., O.K., P.V.V., and J.J.H. analyzed the data; D.D. and F.V.N. performed RNA-seq; N.V., O.K., D.J.C., G.B., P.V.V., and J.J.H. contributed reagents/materials/analysis tools; and S.G., P.V.V. and J.J.H. wrote the manuscript with help from the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jody J. Haigh, Division of Blood Cancers, Australian Centre for Blood Diseases, Department of Clinical Haematology, Monash University and Alfred Health, Alfred Centre, 99 Commercial Rd, Melbourne, VIC 3004, Australia; e-mail: jody.haigh@monash.edu.
References
Author notes
S.G., S.P., P.V.V., and J.J.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal