To the editor:
Genomic loss of the mismatched HLA haplotype (“HLA loss”) represents a frequent mechanism of leukemia-immune evasion and relapse after hematopoietic stem cell transplantation from haploidentical donors (haplo-HSCT).1,2 In the largest retrospective haplo-HSCT series reported to date, HLA loss variants accounted for 23/69 recurrences (33%),3 and their incidence was remarkably similar in other smaller cohorts using either antithymocyte globulin1,4,5 or posttransplant cyclophosphamide6-8 as graft-versus-host-disease (GVHD) prophylaxis. Besides its scientific and epidemiological relevance, documentation of HLA loss has direct and significant clinical implications in informing the choice of salvage treatments because, in this context, the delicate balance between beneficial leukemia control and toxic GVHD associated with donor lymphocyte infusions to treat relapse is heavily shifted toward toxicity because of the loss of the mismatched HLA-haplotype target of alloreactive T cells in HLA loss leukemia cells but not in healthy tissues.2,9,10 Nonetheless, the consensus recommendation to investigate eventual HLA loss at relapse after haplo-HSCT11,12 is difficult to follow for most centers because of methodological and interpretative hurdles, including the frequent need of leukemic blast purification to perform subsequent HLA typing or single nucleotide polymorphism array analysis. In view of the rapid rise in the number of haplo-HSCTs performed worldwide,13,14 the development of user-friendly, yet reliable, sensitive, and cost-effective assays for the detection of HLA loss relapses represents an unmet medical need.
Here we present HLA-KMR, an innovative methodology to detect HLA loss relapses by combination of quantitative polymerase chain reaction (qPCR)-based hematopoietic chimerism, commonly targeting non-HLA gene polymorphisms (non-HLA markers), with ad hoc designed qPCR reactions specific for the most frequent HLA allele groups (HLA markers). The underlying rationale is to use hematopoietic chimerism as a reference to evaluate the presence or absence of the patient-specific HLA. Because the large majority of post-HSCT relapses are of patient origin, they will consequently be positive for patient-specific non-HLA markers. However, whereas “classical” (ie, non-HLA loss) relapses will be concordantly positive also for HLA markers, HLA loss relapses will be positive for non-HLA markers and negative for patient-specific HLA markers because of their selective genomic loss. The comparative evaluation of hematopoietic chimerism with HLA and non-HLA markers could therefore represent an easy tool to discriminate classical from HLA loss relapses (Figure 1A).
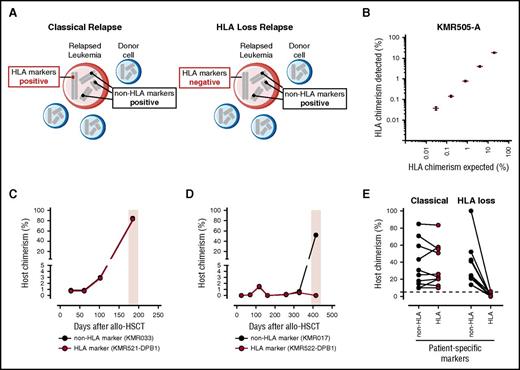
Rationale, validation, and clinical utility of HLA-KMR. (A) Schematic representation of the rationale underlying HLA-KMR. Shown is the simultaneous presence of leukemic blasts (in red) and normal donor hematopoietic cells (in blue) in a typical sample of a patient with relapse after HSCT. In each cell, 4 representative chromosomes are depicted, with a patient-specific HLA marker (in red) and 3 hypothetical non-HLA markers (in black). Leukemic blasts in the classical relapse will be positive for both the HLA and the non-HLA markers (left). Leukemic blasts in the HLA loss relapse will be positive for the non-HLA markers but negative for the HLA markers because of a selective genomic loss of mismatched HLA as an immune escape mechanism of leukemia relapse (right). (B) Validation of the newly developed qPCR assays targeting HLA markers, representatively shown for the KMR505-A assay. Shown are the expected (x-axis) and experimentally determined (y-axis) chimerism measured by serially diluting a target-positive genomic DNA into a target-negative sample. Results from 3 independent experiments are displayed as mean (red dashes) ± standard error of the mean (black whiskers). Data for all other 10 HLA marker-specific qPCR assays developed in this study are reported in supplemental Figure 3. (C-E) Clinical utility of HLA-KMR. (C) Longitudinal posttransplant monitoring by HLA-KMR of a patient who experienced a classical relapse (time of relapse boxed in pink), alongside a patient-specific HLA marker (KMR521-DPB1 reaction, in red) and a patient-specific non-HLA marker (KMRtrack assay KMR033, in black). Note the concordance between the 2 assays at time of relapse, which identifies a classical relapse. (D) Longitudinal posttransplant monitoring by HLA-KMR of a patient who experienced an HLA loss relapse (time of relapse boxed in pink), alongside a patient-specific HLA marker (KMR522-DPB1 reaction, in red) and a patient-specific non-HLA marker (KMRtrack assay KMR017, in black). Note the discordance between the 2 assays at time of relapse, which identifies a HLA loss relapse. (E) Summary of results obtained by chimerism quantification with HLA-KMR on 20 post-HSCT relapses (18 haploidentical, 2 unrelated). As expected, HLA markers (full panel of the newly developed HLA-specific reactions, red dots) and non-HLA markers (KMRtrack assays, black dots) yield concordant results in all classical relapses (left), and discordant results in all HLA loss relapses (right). Because of the relatively low precision of qPCR in measuring high chimerism percentages, quantification of HLA and non-HLA markers in a given sample can have a result slightly different in classical relapses. HLA loss relapses can be unequivocally diagnosed only when HLA markers are negative (<3%, dashed line) and non-HLA markers are positive (>3%).
Rationale, validation, and clinical utility of HLA-KMR. (A) Schematic representation of the rationale underlying HLA-KMR. Shown is the simultaneous presence of leukemic blasts (in red) and normal donor hematopoietic cells (in blue) in a typical sample of a patient with relapse after HSCT. In each cell, 4 representative chromosomes are depicted, with a patient-specific HLA marker (in red) and 3 hypothetical non-HLA markers (in black). Leukemic blasts in the classical relapse will be positive for both the HLA and the non-HLA markers (left). Leukemic blasts in the HLA loss relapse will be positive for the non-HLA markers but negative for the HLA markers because of a selective genomic loss of mismatched HLA as an immune escape mechanism of leukemia relapse (right). (B) Validation of the newly developed qPCR assays targeting HLA markers, representatively shown for the KMR505-A assay. Shown are the expected (x-axis) and experimentally determined (y-axis) chimerism measured by serially diluting a target-positive genomic DNA into a target-negative sample. Results from 3 independent experiments are displayed as mean (red dashes) ± standard error of the mean (black whiskers). Data for all other 10 HLA marker-specific qPCR assays developed in this study are reported in supplemental Figure 3. (C-E) Clinical utility of HLA-KMR. (C) Longitudinal posttransplant monitoring by HLA-KMR of a patient who experienced a classical relapse (time of relapse boxed in pink), alongside a patient-specific HLA marker (KMR521-DPB1 reaction, in red) and a patient-specific non-HLA marker (KMRtrack assay KMR033, in black). Note the concordance between the 2 assays at time of relapse, which identifies a classical relapse. (D) Longitudinal posttransplant monitoring by HLA-KMR of a patient who experienced an HLA loss relapse (time of relapse boxed in pink), alongside a patient-specific HLA marker (KMR522-DPB1 reaction, in red) and a patient-specific non-HLA marker (KMRtrack assay KMR017, in black). Note the discordance between the 2 assays at time of relapse, which identifies a HLA loss relapse. (E) Summary of results obtained by chimerism quantification with HLA-KMR on 20 post-HSCT relapses (18 haploidentical, 2 unrelated). As expected, HLA markers (full panel of the newly developed HLA-specific reactions, red dots) and non-HLA markers (KMRtrack assays, black dots) yield concordant results in all classical relapses (left), and discordant results in all HLA loss relapses (right). Because of the relatively low precision of qPCR in measuring high chimerism percentages, quantification of HLA and non-HLA markers in a given sample can have a result slightly different in classical relapses. HLA loss relapses can be unequivocally diagnosed only when HLA markers are negative (<3%, dashed line) and non-HLA markers are positive (>3%).
For this purpose, we designed and validated a total of 10 qPCR reactions specific for 27 HLA-A, HLA-C, and HLA-DPB1 allele groups, selected based on their frequency in different ethnic populations15 (Table 1; supplemental Table 1, available on the Blood Web site). These 3 loci were chosen because they are often mismatched not only in haplo-HSCT, but also in unrelated donor (UD)-HSCT, a context in which HLA loss relapses have been documented,3,16-18 but in which data on their actual frequency and clinical relevance are missing and warranted. With our qPCR panel, at least 1 informative reaction (ie, targeting at least 1 HLA-A, HLA-C, or HLA-DPB1 allele present in the patient and absent in the donor) was found with a frequency of 70.3% and 66.4%, respectively, in a series of 454 haplo-HSCT and 113 UD-HSCT with a mismatch in the graft-versus-host direction at either of the 3 loci (Table 1).
HLA-specific qPCR assays developed in this study
| Target HLA locus . | Assay name . | Target HLA allele groups† . | Efficiency (%)†,‡ . | Sensitivity (%)†,§ . | Informative HSCT (%)‖ . | |
|---|---|---|---|---|---|---|
| Haploidentical . | Unrelated . | |||||
| (n = 454) . | (n = 113) . | |||||
| A* | KMR501-A | 01, 36 | 87.7 | 0.03 | 10.4 | 0 |
| KMR502-A | 02 | 87.6 | 0.03 | 12.1 | 0 | |
| KMR504-A | 11 | 94.4 | 0.16 | 3.5 | 0.8 | |
| KMR505-A | 23, 24 | 98.0 | 0.03 | 9.9 | 0.8 | |
| KMR506-A | 25, 26, 34, 66, 68, 69 | 94.3 | 0.16 | 11.2 | 4.4 | |
| C* | KMR511-C | 03 | 90.8 | 0.03 | 3.1 | 0 |
| KMR512-C | 04 | 84.5 | 0.16 | 10.6 | 0 | |
| DPB1* | KMR520-DPB1 | 04:01, 23:01, 31:01, 34:01 | 98.3 | 0.03 | 14.1 | 23.8 |
| KMR521-DPB1 | 02:01, 04:02, 08:01, 105:01, 16:01 | 100.8 | 0.03 | 17.2 | 31.8 | |
| KMR522-DPB1 | 01:01, 11:01, 13:01, 15:01 | 96.8 | 0.03 | 3.1 | 16.8 | |
| Target HLA locus . | Assay name . | Target HLA allele groups† . | Efficiency (%)†,‡ . | Sensitivity (%)†,§ . | Informative HSCT (%)‖ . | |
|---|---|---|---|---|---|---|
| Haploidentical . | Unrelated . | |||||
| (n = 454) . | (n = 113) . | |||||
| A* | KMR501-A | 01, 36 | 87.7 | 0.03 | 10.4 | 0 |
| KMR502-A | 02 | 87.6 | 0.03 | 12.1 | 0 | |
| KMR504-A | 11 | 94.4 | 0.16 | 3.5 | 0.8 | |
| KMR505-A | 23, 24 | 98.0 | 0.03 | 9.9 | 0.8 | |
| KMR506-A | 25, 26, 34, 66, 68, 69 | 94.3 | 0.16 | 11.2 | 4.4 | |
| C* | KMR511-C | 03 | 90.8 | 0.03 | 3.1 | 0 |
| KMR512-C | 04 | 84.5 | 0.16 | 10.6 | 0 | |
| DPB1* | KMR520-DPB1 | 04:01, 23:01, 31:01, 34:01 | 98.3 | 0.03 | 14.1 | 23.8 |
| KMR521-DPB1 | 02:01, 04:02, 08:01, 105:01, 16:01 | 100.8 | 0.03 | 17.2 | 31.8 | |
| KMR522-DPB1 | 01:01, 11:01, 13:01, 15:01 | 96.8 | 0.03 | 3.1 | 16.8 | |
Efficiency and sensitivity were tested on HLA-typed B lymphoblastoid cell lines obtained from the European Collection of Animal Cell Cultures or from healthy HLA-typed individuals (supplemental Table 3). For assays targeting multiple HLA allele groups, the most frequent HLA allele group in worldwide populations15 (supplemental Table 1) was used as target-positive reference DNA. Efficiency and sensitivity were determined by serial dilutions of target-positive reference DNA in water.
Reported percentages are the mean of at least 3 independent experiments performed at either of the 2 testing sites (Ospedale San Raffaele [OSR] and Universitätsklinikum Essen [UKE]). Standard error of mean was <4 in all cases.
Maximal reproducible sensitivity determined in at least 3 independent experiments performed at either of the 2 testing sites (OSR and UKE).
Percentage of informative HSCT based on HLA typing of patients and donors for 454 haplo-HSCT performed between years 2000 and 2016 at OSR, and for 113 UD-HSCT performed in 2015 at UKE with at least 1 HLA-A, HLA-C, or HLA-DPB1 mismatch in the graft-versus-host direction. Some transplants had more than 1 informative reaction, whereas others had none. The total percentages of transplants with at least 1 informative reaction was 71.6% and 66.4% for haplo-HSCT and UD-HSCT, respectively.
To validate their technical robustness and reproducibility, the HLA-specific reactions were extensively tested at 2 independent sites (OSR and UKE) on different combinations of HLA-typed reference DNAs. Details on the experimental methods are in the supplemental Data. On-target amplification efficiency averaged to 92.8% (Table 1), with superimposable performance in water and in target-negative DNA (overall concordance by Pearson correlation R2 = 0.99, P < .0001; supplemental Figure 1). Specificity and possible cross-reactivity were tested both in silico based on nucleotide sequences reported in the Immunogene Tics/HLA database19 (supplemental Table 2) and in a panel of 45 HLA-typed reference DNAs (supplemental Table 3), demonstrating high specificity for the target HLA allele groups (supplemental Figure 2). Importantly, accuracy and precision in chimerism determination resulted very high for all reactions, with almost perfect concordance between the expected and experimentally determined quantification in serial dilutions of HLA target-positive into target-negative DNA (overall concordance by Pearson correlation R2 = 0.99, P < .0001; shown as representative for reaction KMR505-A in Figure 1B and for all reactions in supplemental Figure 3). Maximal reproducible sensitivity defined according to minimum information for publication of quantitative real-time PCR experiments guidelines20 was at least 0.16% for all reactions.
The clinical utility of the newly developed assays was tested by analyzing a total of 20 cases of leukemia relapse after haplo-HSCT or UD-HSCT (for details, see supplemental Data). These relapses had been previously shown by single nucleotide polymorphism arrays (Illumina Human CNV370-Quad BeadArray or Human660W-Quad BeadChip, Illumina) or sequence-specific oligonucleotide hybridization HLA typing (INNO-LiPA, Fujirebio Inc.)1,3,16 to be classical (n = 10) or HLA loss (n = 10). We reanalyzed these relapses by combining our newly developed HLA-specific assays with commercial qPCR chimerism assays targeting 39 non-HLA biallelic variant markers (KMRtype and KMRtrack assays, GenDx, Utrecht, The Netherlands). Figure 1C-D present as an example data obtained for 2 representative patients with a classical and an HLA loss relapse, respectively. Both cases showed concordant low (<3%) positivity for HLA and non-HLA markers in samples obtained prior to relapse, suggesting the detection of residual HLA-heterozygous host chimerism, in line with our previous data on qPCR chimerism monitoring21 and with the results obtained from 3 patients with long-term complete remission analyzed with HLA-KMR (supplemental Figure 4). At the time of classical relapse, positivity for both HLA and non-HLA markers rose to similar levels, both consistent with the blast percentage detected at bone marrow examination (data not shown). In contrast, the HLA loss relapse sample showed positivity for the non-HLA marker that was again in line with the morphological blast percentage (data not shown), but as expected the inside HLA reaction marker remained negative (<1%). Similar data were obtained for all 10 classical and 10 HLA relapse cases analyzed (Figure 1E). Of note, to cover the 20 cases, each of the 10 newly developed HLA-specific qPCR assays was employed at least once, documenting the relevance of developing multiple HLA markers and the good coverage of our panel.
Our HLA-KMR methodology for rapid and reliable univocal discrimination of HLA loss from classical relapse fills a gap in the scientific and diagnostic follow-up of allogeneic HSCT. First, it provides an economic and user-friendly method applicable at all centers performing haplo-HSCT. In view of the exponential increase in the haploidentical transplant activity registered worldwide in the past few years,13,14 this is of interest to a growing number of centers to decide on whether to perform or withhold donor lymphocyte infusions. Given the high sensitivity of the assays, HLA-KMR might even enable the detection of imminent HLA loss relapse before its actual onset, although these relapses are known to occur late after HSCT,1,3 when diagnostic monitoring tends to thin out. Second, HLA-KMR will facilitate retrospective and prospective studies apt to understand the actual frequency and potential clinical relevance of HLA loss relapses in adult volunteer UD-HSCT, the most frequent allogeneic transplant setting to date,13 as well as in umbilical cord blood-HSCT.
Although the qPCR reactions in the HLA-KMR system do not cover all HLA-A, HLA-C, and HLA-DPB1 allele groups, they were informative in more than two-thirds of potentially informative HSCT from haploidentical or unrelated donors. Complete HLA allele coverage could be offered by next-generation sequencing approaches that are being developed for chimerism.22,23 Moreover, the relatively low precision of qPCR especially to quantify high chimerism percentages, could be improved in the future by use of digital PCR.24,25 We deliberately chose a qPCR platform to provide a methodology at immediate reach to transplant centers throughout the world, including those with limited technological and economical resources that are among the most rapidly increasing clinical users of haplo-HSCT. For all of these centers, the HLA-KMR approach enables discrimination between HLA loss and classical relapse in an affordable manner and in less than working day, thus facilitating rapid clinical decision-making and, ultimately, patient-tailored therapeutic interventions.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: This work was supported by the European Commission Transcan JTC2012 (Cancer12-045-HLALOSS) (L.V., D.W.B., and K.F.), Deutsche José Carreras Leukämie Stiftung (DJCLS R 15/02) (D.W.B. and K.F.), Werner Jackstädt Stiftung (K.F.), Joseph-Senker Stiftung (K.F.), Italian Ministry of Health RF-2011-02351998 (F.C. and L.V.), RF-2011-02348034 (L.V.), Associazione Italiana per la Ricerca sul Cancro Start-Up Grant #14162 (L.V.), Conquer Cancer Foundation 2014 Young Investigator Award (L.V.), and DKMS Mechtild Harf Research Grant (L.V.). C.T. was supported by a postdoctoral fellowship from the Associazione Italiana per la Ricerca sul Cancro.
Contribution: M.A., C.T., K.F., and L.V. designed the study; M.A., C.T., E.B., P.C., C.B., C.P., K.S., D.B., and B.M. performed and analyzed the experiments; D.W.B., F.C., and L.V. contributed to patient clinical care and data collection; W.M. provided scientific advice and supervision; M.A., C.T., K.F., and L.V. wrote the paper; and all authors read and approved the final version of the manuscript.
Conflict-of-interest disclosure: E.B. and W.M. are employees of GenDx, Utrecht, The Netherlands. K.F. and L.V. received research funding from GenDx, Utrecht, The Netherlands. The remaining authors declare no competing financial interests.
Correspondence: Katharina Fleischhauer, Institute for Experimental Cellular Therapy, University Hospital Essen, Hufelandstrasse 55, 45147 Essen, Germany; e-mail: katharina.fleischhauer@uk-essen.de; or Luca Vago, Unit of Immunogenetics, Leukemia Genomics and Immunobiology, IRCCS San Raffaele Scientific Institute, Via Olgettina 60, 20132 Milan, Italy; e-mail: vago.luca@hsr.it.
References
Author notes
M.A. and C.T. contributed equally to this study.
K.F. and L.V. contributed equally to this study.