Key Points
Variation in the Fcγ receptor gene cluster is associated with protection from RBC alloimmunization in patients with SCD.
This association appears to be strongest for alloimmunization to antigens other than the immunogenic Rh or K.
Abstract
Red blood cell (RBC) transfusions are of vital importance in patients with sickle cell disease (SCD). However, a major complication of transfusion therapy is alloimmunization. The low-affinity Fcγ receptors, expressed on immune cells, are important regulators of antibody responses. Genetic variation in FCGR genes has been associated with various auto- and alloimmune diseases. The aim of this study was to evaluate the association between genetic variation of FCGR and RBC alloimmunization in SCD. In this case-control study, DNA samples from 2 cohorts of transfused SCD patients were combined (France and The Netherlands). Cases had a positive history of alloimmunization, having received ≥1 RBC unit. Controls had a negative history of alloimmunization, having received ≥20 RBC units. Single nucleotide polymorphisms and copy number variation of the FCGR2/3 gene cluster were studied in a FCGR-specific multiplex ligation-dependent probe amplification assay. Frequencies were compared using logistic regression. Two hundred seventy-two patients were included (130 controls, 142 cases). The nonclassical open reading frame in the FCGR2C gene (FCGR2C.nc-ORF) was strongly associated with a decreased alloimmunization risk (odds ratio [OR] 0.26, 95% confidence [CI] 0.11-0.64). This association persisted when only including controls with exposure to ≥100 units (OR 0.30, CI 0.11-0.85) and appeared even stronger when excluding cases with Rh or K antibodies only (OR 0.19, CI 0.06-0.59). In conclusion, SCD patients with the FCGR2C.nc-ORF polymorphism have over a 3-fold lower risk for RBC alloimmunization in comparison with patients without this mutation. This protective effect was strongest for exposure to antigens other than the immunogenic Rh or K antigens.
Introduction
Red blood cell (RBC) transfusions are of vital importance in patients with sickle cell disease (SCD). However, an important and frequent complication of transfusion therapy is alloimmunization.1 The development of alloantibodies against foreign RBC antigens complicates donor-matching procedures and significantly limits the availability of matching blood for future transfusions. In addition, alloimmunized patients are at risk of developing delayed hemolytic transfusion reactions (DHTR), a potentially lethal complication that is often characterized by hemoglobinuria and severe vaso-occlusive crisis (VOC) with a high frequency of secondary acute chest syndrome.2,3
Alloimmunization occurs significantly more frequently in SCD patients than in the general population, with incidences ranging from 18% to 76% with ABO and Rhesus D antigen matching only.4,5 This high alloimmunization rate in SCD patients can partly be explained by the differences in RBC antigens between donors, primarily of European descent, and recipients, primarily of African descent.6 Extended antigen matching for Rhesus (Rh) phenotype and Kell (K) has significantly reduced the rate of alloimmunization, whereas additional antigen matching for Fy, Jk, and MNS appears to be even more effective.7-12 Unfortunately, extended matching is not always applied because of limited availability of matching units and the high costs involved.12 Moreover, phenotypic matching does not always prevent alloimmunization against these highly immunogenic antigens.13,14 A study by Chou et al demonstrated that 58% of chronically transfused SCD patients were alloimmunized despite phenotypic matching for Rh phenotype and K, because of the high genetic variation in Rh genes in both the SCD population and the African American donors, as were used in this study.14
Interestingly, only a subgroup of patients appears to develop alloantibodies after transfusion.4,15,16 It is yet unclear why some patients develop alloantibodies (responders), whereas others remain tolerant despite high transfusion exposures (nonresponders). A better understanding of the pathophysiology of RBC alloimmunization may help to identify these high-risk patients and could add to the detection of targets for preventive strategies, ultimately promoting a safer and more cost-effective application of transfusion therapy in SCD.17
The basic concept of alloimmunization involves the uptake of transfused allogenic RBC antigens by antigen-presenting cells, presentation of antigen by the HLA class II complex to CD4+ T helper cells, and subsequent B-cell activation with antibody production.18,19 However, the occurrence of an actual antibody response is determined by a complex interplay of both genetic predisposition and circumstantial factors at time of transfusion, such as the extent of antigenic incompatibility between donor and host, the immunogenicity of a specific alloantigen, and the level of systemic inflammation.1,20-23 As for genetic predisposition, the HLA class II genotype of a patient is a potential predictor of the alloimmunization status, in SCD populations as in general transfused populations.20,24-28 In addition, polymorphisms in immunoregulatory genes have been implicated in RBC alloimmunization (TRIM21, CD81).29,30
This is the first study to assess the association between RBC alloimmunization in SCD and the genes encoding for low-affinity Fcγ receptors (FcγRs) for immunoglobin G (IgG). FcγRs are glycoproteins, expressed mainly by immune effector cells such as antigen-presenting cells, macrophages, and monocytes. They allow these cells to bind to the Fc portion of IgG, attached to circulating antigen, facilitating activation of the cell or presentation of antigenic peptides. FcγRIIb and FcγRIII are believed to mediate antigen internalization and presentation to T cells.31 Evidence supporting this importance of FcγRs in antigen uptake and presentation is provided in studies that show that a single amino acid mutation in the FcγRIIb or FcγRIII cytoplasmic tail prevents antigen internalization and presentation.32 FcγRIIb appears to play an important role in the regulation of B cells and antibody-producing plasma cells.33 The receptor has been proposed to maintain peripheral tolerance for B cells,34 a theory that is supported by the fact that FcγRIIb knockout mice lose peripheral B-cell tolerance, produce autoantibodies, and develop autoimmune disease.35-37 Furthermore, FcγRIIb regulates affinity maturation and memory B-cell development.38,39 Moreover, FcγRs can shape the antibody repertoire by modulating B-cell receptor-mediated cell activation and proliferation.34,40 These effects emphasize the crucial role of FcγRs in the formation of alloantibodies.
The family of FcγRs includes the high-affinity FcγRI and the low-affinity FcγRII and FcγRIII, comprising various subclasses (FcγRIIa, FcγRIIb, FcγRIIc, FcγRIIIa, and FcγRIIIb). These low-affinity FcγRs provide both pro- and anti-inflammatory regulation of immune responses and are encoded by the genes FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B.33 Polymorphisms in these genes are common and may affect the function of these proteins, thereby affecting the balance between activating and inhibitory signaling pathways. Various polymorphisms in FCGR genes have been associated with the occurrence of certain auto- and alloimmune diseases, such as idiopathic thrombocytopenic purpura, systemic lupus erythematosus, and inhibitor development in hemophilia A.41-44
The aim of this study was to evaluate whether polymorphisms in the FCGR2 and FCGR3 genes are associated with RBC alloimmunization in a cohort of SCD patients. A secondary aim of this study was to assess whether variation within these genes confers susceptibility for the occurrence of a DHTR.
Methods
A detailed description and additional methods are available in the supplemental Methods, available on the Blood Web site.
Patients
For this observational, case-control study, we combined 2 longitudinal cohorts of transfused SCD patients with retrospective data on transfusion exposure and alloimmunization status: one from The Netherlands (STAR cohort, N = 342)4 and one from France (SCDTRANSFU cohort, N = 250, including 75 identified DHTR cases3 ). Patients from both cohorts were included in this study if (a) a DNA sample was available, and (b) they had a negative history of alloimmunization with a minimum transfusion exposure of ≥20 RBC units (controls) or a positive history of alloimmunization with a minimum transfusion exposure of ≥1 RBC unit (cases). A minimum of 20 units was applied in negative patients to assure that patients had sufficient transfusion exposure to potentially form any alloantibodies (nonresponders).4 In addition, because there was a maximum number of samples agreed upon to be shipped for the French cohort, a small fraction of patients from the French source population were excluded, on the basis of their chronological entry in the French dataset.
The institutional review boards of all participating centers approved the study. This study was conducted in accordance with the Declaration of Helsinki.
Data collection and definitions
In the primary analysis of this study, we assessed the association between polymorphisms in the FCGR2 and FCGR3 genes and RBC alloimmunization. Cases were defined here as all patients with a clinically significant alloantibody. Controls were defined as all patients with a negative history of alloimmunization, with a minimum transfusion exposure of ≥20 RBC units. Patients with exclusively 1 of the following antibodies were excluded from this analysis: autoantibodies, cold antibodies, and naturally occurring antibodies such as anti-A1, anti-IH, anti-H, anti-A1, anti-I, anti-Lea, anti-Leb, and anti-P1. Anti-M was defined as naturally occurring if demonstrated upon first antibody screening with no history of prior transfusion exposure. Transfusion and laboratory policies can be found in the supplemental Methods.
To test the robustness of our findings, we performed a sensitivity analysis using a more strict definition for nonresponders, excluding controls with <100 units of transfusion exposure. In addition, a second sensitivity analysis was performed to assess the effect of immunization against the highly immunogenic Rh and K antigens (RhD, C, c, E, e, and K) on the association between FCGR polymorphisms and alloimmunization. We hypothesized that the genetic constitution of a patient is less important for the formation of Rh- and K-specific alloantibodies because these antibodies are so readily formed.20,21,45 To test this hypothesis, we categorized alloimmunized patients (cases) into 2 subgroups: the first group contained patients with exclusively Rh- or K-specific antibodies; the second group contained patients with at least 1 antibody other than Rh or K. Associations between FCGR polymorphisms and alloimmunization were assessed and compared between these 2 subgroups.
Lastly, we performed a separate analysis in the French cohort to assess associations between FCGR polymorphisms and the occurrence of DHTR. DHTR information was solely available for patients from the French cohort. In this analysis, cases were defined as all patients with a positive history of DHTR. Controls were defined as patients with a negative history of DHTR, with a minimum transfusion exposure of at least 20 RBC units, regardless of the alloimmunization status. DHTR was defined by signs of accelerated hemolysis (including hemoglobinuria, lactic dehydrogenase), with a more severe anemia than was present before transfusion, and development of symptoms suggestive of a VOC or acute chest syndrome, appearing ≥3 days after transfusion.46
Statistical analysis
The allele frequencies of single nucleotide polymorphisms (SNPs) and copy number variation (CNV) were compared between alloimmunized and nonalloimmunized patients and between DHTR and non-DHTR patients, using single variant logistic regression, adjusted for cohort (French or Dutch) as a covariate, and assuming an additive genetic effect (in case of variants with more than 2 genotypes). Comparisons were expressed in odds ratios (OR), 95% confidence intervals (CI), and corresponding P values. In our primary analysis, we applied a Bonferroni correction to adjust for multiple testing. A P value of <.0038 (0.05 of 13 variants) was considered statistically significant here. In all subsequent sensitivity analyses, a P value of <.05 for significance was applied. Expression levels of FcγRs were analyzed using GraphPad Prism 7.02. For comparison of median fluorescence intensity (MFI) the 1-way analysis of variance was used, followed by Sidak post hoc test for correction of multiple comparisons (P < .0001; P < .001; P < .01; P < .05).
Results
Study population
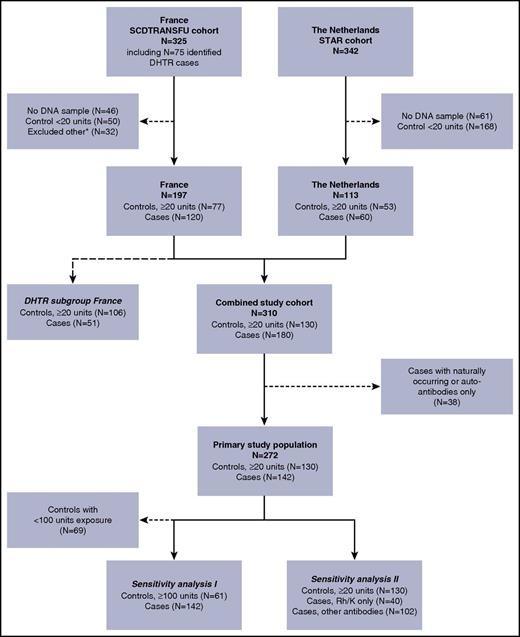
Our source study population comprised transfused SCD patients from a cohort from France (N = 325) and a cohort from the Netherlands (N = 342; Figure 1). Of these, 272 patients (130 controls and 142 alloimmunized cases) were eligible for this study and formed our primary study population. Baseline characteristics of this primary study population are described in Table 1. Patients in the French cohort were slightly older, had a higher cumulative transfusion exposure, and had a higher proportion of patients of African ethnicity than did those in the Dutch cohort, which consisted mostly of patients of Latin American origin (mainly Surinam and former Dutch Caribbean islands). An overview of all clinically significant alloantibodies and the number of alloantibodies per patient are demonstrated in supplemental Tables 1 and 2, respectively.
Flowchart of patients included in the study. Our source study population comprised transfused SCD patients from a cohort from France (FR; N = 325), and a cohort from the Netherlands (NL; N = 342). Of these, 310 patients were eligible for this study (DNA sample available, alloimmunized cases or controls with ≥20 units exposure). An additional 38 cases were excluded because they exclusively had either autoantibodies or naturally occurring antibodies (patients excluded: FR N = 29, NL N = 9). The remaining 272 patients (130 controls and 142 alloimmunized cases) formed our primary study population. We performed 2 sensitivity analyses: (I) excluding controls with <100 units of transfusion exposure (patients excluded: FR N = 36, NL N = 33) and (II) dividing cases into patients who had only antibodies with Rh or K specificity and patients with at least 1 antibody other than Rh or K. A separate analysis was performed to assess the association of FCGR polymorphisms with the occurrence of DHTR. Data on the history of DHTR status was available only in a subset of the FR cohort (N = 157; 28 patients were excluded because of unknown DHTR status, and 12 controls were excluded because they had <20 units of exposure). *Excluded other: because there was a maximum number of samples of the French cohort agreed upon to be shipped, a small fraction of patients from the French source population were excluded on the basis of their chronological entry in the dataset.
Flowchart of patients included in the study. Our source study population comprised transfused SCD patients from a cohort from France (FR; N = 325), and a cohort from the Netherlands (NL; N = 342). Of these, 310 patients were eligible for this study (DNA sample available, alloimmunized cases or controls with ≥20 units exposure). An additional 38 cases were excluded because they exclusively had either autoantibodies or naturally occurring antibodies (patients excluded: FR N = 29, NL N = 9). The remaining 272 patients (130 controls and 142 alloimmunized cases) formed our primary study population. We performed 2 sensitivity analyses: (I) excluding controls with <100 units of transfusion exposure (patients excluded: FR N = 36, NL N = 33) and (II) dividing cases into patients who had only antibodies with Rh or K specificity and patients with at least 1 antibody other than Rh or K. A separate analysis was performed to assess the association of FCGR polymorphisms with the occurrence of DHTR. Data on the history of DHTR status was available only in a subset of the FR cohort (N = 157; 28 patients were excluded because of unknown DHTR status, and 12 controls were excluded because they had <20 units of exposure). *Excluded other: because there was a maximum number of samples of the French cohort agreed upon to be shipped, a small fraction of patients from the French source population were excluded on the basis of their chronological entry in the dataset.
Patient characteristics for the primary study population (N = 272)
| N (%) or median (IQR) . | France (N = 168) . | The Netherlands (N = 104) . |
|---|---|---|
| Age at last follow-up in years | 32 (27-40) | 28 (21-38) |
| Female sex | 91 (54) | 62 (60) |
| Hemoglobin genotype | ||
| HbSS/HbSβ0 | 163 (97) | 91 (88) |
| HbSC/HbSβ+ | 5 (3) | 13 (12) |
| Ethnicity* | ||
| Africa | 143 (85) | 26 (25) |
| Latin America | 18 (11) | 71 (68) |
| Asia | 0 (0) | 3 (3) |
| Other | 2 (1) | 4 (4) |
| Cumulative transfusion exposure, units | 72 (13-179) | 27 (9-77) |
| N (%) or median (IQR) . | France (N = 168) . | The Netherlands (N = 104) . |
|---|---|---|
| Age at last follow-up in years | 32 (27-40) | 28 (21-38) |
| Female sex | 91 (54) | 62 (60) |
| Hemoglobin genotype | ||
| HbSS/HbSβ0 | 163 (97) | 91 (88) |
| HbSC/HbSβ+ | 5 (3) | 13 (12) |
| Ethnicity* | ||
| Africa | 143 (85) | 26 (25) |
| Latin America | 18 (11) | 71 (68) |
| Asia | 0 (0) | 3 (3) |
| Other | 2 (1) | 4 (4) |
| Cumulative transfusion exposure, units | 72 (13-179) | 27 (9-77) |
Ethnicity was unknown in 5 patients in the French cohort (N = 163).
FCGR gene CNV or polymorphism and susceptibility to alloimmunization
We have analyzed the low-affinity FCGR2/3 genes of SCD patients with or without alloimmunization by genotyping CNV and SNPs by multiplex ligation-dependent probe amplification (MLPA).
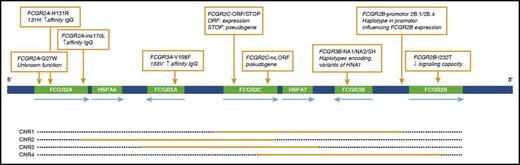
The FCGR2/3 gene locus contains 4 distinct CNV regions (CNR),47 as are shown in Figure 2. Table 2 shows the frequencies of the tested CNRs. CNV was found most frequently in CNR1 (FCGR2C/FCGR3B), for which 56 patients showed a change in copy number. Copy number changes in the 4 CNRs were not significantly associated with susceptibility to alloimmunization in our study population.
Schematic overview of the FCGR2/3 locus. Nine SNPs and haplotypes are indicated with orange boxes. Orange bars depict the approximate extent of CNRs in which duplication or deletion can occur. HNA, human neutrophil antigen.
Schematic overview of the FCGR2/3 locus. Nine SNPs and haplotypes are indicated with orange boxes. Orange bars depict the approximate extent of CNRs in which duplication or deletion can occur. HNA, human neutrophil antigen.
Copy numbers and allele frequencies of FCGR2/3 genetic variation in 130 control patients (>20 transfused units, no alloantibodies) and 142 alloimmunized patients
| . | Controls/no alloimmunization, n (%) . | Cases/alloimmunized patients, n (%) . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 1 . | 2 . | 3 . | 4 . | 0 . | 1 . | 2 . | 3 . | 4 . | OR . | 95% CI . | P . |
| Copy number region | |||||||||||||
| CNR1 | 0 (0) | 15 (12) | 105 (81) | 9 (7) | 1 (1) | 3 (2) | 18 (13) | 111 (78) | 10 (7) | 0 (0) | 0.76 | 0.47-1.24 | .271 |
| CNR2 | 0 (0) | 1 (1) | 128 (98) | 1 (1) | 0 (0) | 0 (0) | 2 (1) | 137 (96) | 3 (2) | 0 (0) | 1.27 | 0.28-5.68 | .758 |
| CNR3 | 0 (0) | 0 (0) | 128 (99) | 1 (1) | 0 (0) | 0 (0) | 1 (1) | 138 (97) | 3 (2) | 0 (0) | 1.55 | 0.25-9.44 | .635 |
| CNR4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | na | na | na |
| Allele frequencies | |||||||||||||
| FCGR2A-131H | 45 (35) | 62 (48) | 23 (18) | 0 (0) | 0 (0) | 43 (30) | 77 (54) | 22 (15) | 0 (0) | 0 (0) | 1.27 | 0.76-2.14 | .358 |
| FCGR2A-27W | 112 (86) | 18 (14) | 0 (0) | 0 (0) | 0 (0) | 128 (90) | 14 (10) | 0 (0) | 0 (0) | 0 (0) | 0.69 | 0.33-1.46 | .334 |
| FCGR3A-158V | 53 (41) | 59 (45) | 18 (14) | 0 (0) | 0 (0) | 66 (46) | 56 (39) | 20 (14) | 0 (0) | 0 (0) | 0.91 | 0.65-1.29 | .598 |
| FCGR2C-ORF | 125 (96) | 5 (4) | 0 (0) | 0 (0) | 0 (0) | 138 (97) | 4 (3) | 0 (0) | 0 (0) | 0 (0) | 0.75 | 0.20-2.87 | .675 |
| FCGR2C.nc-ORF | 109 (84) | 21 (16) | 0 (0) | 0 (0) | 0 (0) | 135 (95) | 7 (5) | 0 (0) | 0 (0) | 0 (0) | 0.26 | 0.11-0.64 | .003 |
| FCGR3B-NA1 | 44 (34) | 63 (49) | 22 (17) | 0 (0) | 0 (0) | 58 (41) | 60 (43) | 21 (15) | 2 (1) | 0 (0) | 0.89 | 0.64-1.23 | .471 |
| FCGR3B-SH | 93 (72) | 35 (27) | 2 (2) | 0 (0) | 0 (0) | 102 (72) | 34 (24) | 6 (4) | 0 (0) | 0 (0) | 1.07 | 0.68-1.69 | .772 |
| FCGR2B-232T | 71 (55) | 53 (41) | 6 (5) | 0 (0) | 0 (0) | 72 (51) | 57 (40) | 13 (9) | 0 (0) | 0 (0) | 1.16 | 0.72-1.87 | .537 |
| FCGR2B haplotype 2B.4 | 127 (98) | 3 (2) | 0 (0) | 0 (0) | 0 (0) | 139 (99) | 2 (1) | 0 (0) | 0 (0) | 0 (0) | 0.64 | 0.10-3.91 | .628 |
| . | Controls/no alloimmunization, n (%) . | Cases/alloimmunized patients, n (%) . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 1 . | 2 . | 3 . | 4 . | 0 . | 1 . | 2 . | 3 . | 4 . | OR . | 95% CI . | P . |
| Copy number region | |||||||||||||
| CNR1 | 0 (0) | 15 (12) | 105 (81) | 9 (7) | 1 (1) | 3 (2) | 18 (13) | 111 (78) | 10 (7) | 0 (0) | 0.76 | 0.47-1.24 | .271 |
| CNR2 | 0 (0) | 1 (1) | 128 (98) | 1 (1) | 0 (0) | 0 (0) | 2 (1) | 137 (96) | 3 (2) | 0 (0) | 1.27 | 0.28-5.68 | .758 |
| CNR3 | 0 (0) | 0 (0) | 128 (99) | 1 (1) | 0 (0) | 0 (0) | 1 (1) | 138 (97) | 3 (2) | 0 (0) | 1.55 | 0.25-9.44 | .635 |
| CNR4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | na | na | na |
| Allele frequencies | |||||||||||||
| FCGR2A-131H | 45 (35) | 62 (48) | 23 (18) | 0 (0) | 0 (0) | 43 (30) | 77 (54) | 22 (15) | 0 (0) | 0 (0) | 1.27 | 0.76-2.14 | .358 |
| FCGR2A-27W | 112 (86) | 18 (14) | 0 (0) | 0 (0) | 0 (0) | 128 (90) | 14 (10) | 0 (0) | 0 (0) | 0 (0) | 0.69 | 0.33-1.46 | .334 |
| FCGR3A-158V | 53 (41) | 59 (45) | 18 (14) | 0 (0) | 0 (0) | 66 (46) | 56 (39) | 20 (14) | 0 (0) | 0 (0) | 0.91 | 0.65-1.29 | .598 |
| FCGR2C-ORF | 125 (96) | 5 (4) | 0 (0) | 0 (0) | 0 (0) | 138 (97) | 4 (3) | 0 (0) | 0 (0) | 0 (0) | 0.75 | 0.20-2.87 | .675 |
| FCGR2C.nc-ORF | 109 (84) | 21 (16) | 0 (0) | 0 (0) | 0 (0) | 135 (95) | 7 (5) | 0 (0) | 0 (0) | 0 (0) | 0.26 | 0.11-0.64 | .003 |
| FCGR3B-NA1 | 44 (34) | 63 (49) | 22 (17) | 0 (0) | 0 (0) | 58 (41) | 60 (43) | 21 (15) | 2 (1) | 0 (0) | 0.89 | 0.64-1.23 | .471 |
| FCGR3B-SH | 93 (72) | 35 (27) | 2 (2) | 0 (0) | 0 (0) | 102 (72) | 34 (24) | 6 (4) | 0 (0) | 0 (0) | 1.07 | 0.68-1.69 | .772 |
| FCGR2B-232T | 71 (55) | 53 (41) | 6 (5) | 0 (0) | 0 (0) | 72 (51) | 57 (40) | 13 (9) | 0 (0) | 0 (0) | 1.16 | 0.72-1.87 | .537 |
| FCGR2B haplotype 2B.4 | 127 (98) | 3 (2) | 0 (0) | 0 (0) | 0 (0) | 139 (99) | 2 (1) | 0 (0) | 0 (0) | 0 (0) | 0.64 | 0.10-3.91 | .628 |
Entries in boldface type indicate significant values.
na, not applicable.
In addition, polymorphisms in the FCGR2/3 locus were analyzed for association with alloimmunization (Table 2). Single-variant logistic regression revealed a highly significant association for the FCGR2C nonclassical open reading frame haplotype (FCGR2C.nc-ORF) (OR, 0.26; CI, 0.11-0.64; P = .003). This OR indicates a protective effect of this polymorphism for alloimmunization. Only 7 of the 142 (5%) alloimmunized patients had this polymorphism, in comparison with 21 of the 130 (16%) nonimmunized control patients. This association was consistent in our sensitivity analysis when including only controls with a transfusion exposure of ≥100 units (Figure 1; OR, 0.30; CI, 0.11-0.85; P = .023). Other polymorphisms were not associated with alloimmunization.
Interestingly, when excluding cases with antibodies exclusively against the relatively immunogenic Rh and K antigens, the protective association of the FCGR2C.nc-ORF polymorphism with alloimmunization appeared to be even stronger (Figure 1; Table 3; OR, 0.19; CI, 0.06-0.59; P = .004). In contrast, when including only these cases with Rh- or K- specific antibodies in our analysis, the association was not significant anymore (OR, 0.50; CI, 0.14-1.81; P = .292).
The influence of highly immunogenic Rhesus or Kell alloantibodies on the association between the FCGR2C.nc-ORF polymorphism and alloimmunization
| . | OR . | CI . | P . | N (control/cases) . |
|---|---|---|---|---|
| Controls vs alloimmunized cases, cases, exclusively Rh or K antibodies* | ||||
| FCGR2C.nc-ORF | 0.50 | 0.14-1.81 | .292 | 130/40 |
| Controls vs alloimmunized cases, cases, at least 1 antibody other than Rh or K | ||||
| FCGR2C.nc-ORF | 0.19 | 0.06-0.59 | .004 | 130/102 |
| . | OR . | CI . | P . | N (control/cases) . |
|---|---|---|---|---|
| Controls vs alloimmunized cases, cases, exclusively Rh or K antibodies* | ||||
| FCGR2C.nc-ORF | 0.50 | 0.14-1.81 | .292 | 130/40 |
| Controls vs alloimmunized cases, cases, at least 1 antibody other than Rh or K | ||||
| FCGR2C.nc-ORF | 0.19 | 0.06-0.59 | .004 | 130/102 |
Entry in boldface type indicates a significant value.
RhD, C, c, E, e, or K antibodies.
Despite the clear protective association, 7 patients with the FCGR2C.nc-ORF polymorphism in the total study population still formed alloantibodies. Table 4 demonstrates that of these 7 patients, 3 patients developed only antibodies with Rh specificity within 1-9 units of RBC transfusions. Two patients primarily developed a Rh antibody and subsequently formed additional alloantibodies. The remaining 2 patients developed anti-Jsa (KEL 6) or anti-Kpa (KEL 3) antibodies, which both belong to the Kell antibody system.48 These are both low-incidence antigens, and therefore the likelihood of exposure to the antigens by transfusion is low.
Patient data showing specificity of antibodies
| Alloimmunized cases FGCR2C.nc-ORF . | Antibody . | Units to first antibody . |
|---|---|---|
| Patient nc-1 | Anti-C | 1 |
| Patient nc-2 | Anti-Jsa | 31 |
| Patient nc-3 | Anti-E | 2 |
| Patient nc-4 | Anti-E | 9 |
| Patient nc-5 | Primary: Anti-E/M/Jkb | >3 |
| Following: anti-S, anti-Fya, anti-H | ||
| Patient nc-6 | Anti-Kpa | 16 |
| Patient nc-7 | Primary: Anti-E | 2 |
| Following: anti-Knops- auto (RH4-RH6-RH5)- FY5-MNS1-MNS3-LE1-LE2-YT2-Htl-ASP |
| Alloimmunized cases FGCR2C.nc-ORF . | Antibody . | Units to first antibody . |
|---|---|---|
| Patient nc-1 | Anti-C | 1 |
| Patient nc-2 | Anti-Jsa | 31 |
| Patient nc-3 | Anti-E | 2 |
| Patient nc-4 | Anti-E | 9 |
| Patient nc-5 | Primary: Anti-E/M/Jkb | >3 |
| Following: anti-S, anti-Fya, anti-H | ||
| Patient nc-6 | Anti-Kpa | 16 |
| Patient nc-7 | Primary: Anti-E | 2 |
| Following: anti-Knops- auto (RH4-RH6-RH5)- FY5-MNS1-MNS3-LE1-LE2-YT2-Htl-ASP |
Lastly, we also evaluated whether the association was consistent in both the French and the Dutch subcohorts of patients. In the French subcohort, the association was strongly present (N = 165; OR, 0.13; CI, 0.04-0.47; P = .002), whereas the association did not persist in Dutch patients (N = 104; OR, 0.82; CI, 0.21-3.23; P = .77). Yet patients in the Dutch subcohort had a higher exposure to units matched only for AB0 and RhD antigens, because the study observation window of this cohort was wider. Therefore, the proportion of cases with exclusively Rh or K antibodies was significantly higher in the Dutch subcohort than in the French subcohort (26 out of 51 cases [51%] and 14 out of 91 cases [15%], respectively; P < .001).
Overall, these data indicate that the protective effect of FCGR2C.nc-ORF appears to be strongest for alloimmunization against antigens other than Rh or K and is therefore most relevant if extended matching for Rh and K antigens is performed.
FCGR gene CNV and SNPs and susceptibility to DHTR
An important clinical complication of alloimmunization is DHTR. Besides their role in the formation of alloantibodies, FcγRs are especially known for their capacity to induce antibody-mediated phagocytosis or antibody-mediated cellular toxicity (ADCC) associated with hemolysis once an alloantibody is present.49 Therefore, we have analyzed the low-affinity FcγR genes of SCD patients to investigate the presence of a potential association with DHTR after alloimmunization. The history of DHTR status was available in 157 patients of the French cohort (Figure 1). One hundred and six of these patients did not have a history of DHTR and served as controls. Fifty-one patients did develop DHTR and were defined as cases. Although a trend of association was found again for FCGR2C.nc-ORF (OR, 0.48; CI, 0.15-1.51; P = .210), single-variant logistic regression analysis did not show any significant associations between FcγR polymorphisms or CNV and DHTR (supplemental Table 3).
Functional consequence of the FCGR2C.nc-ORF polymorphism
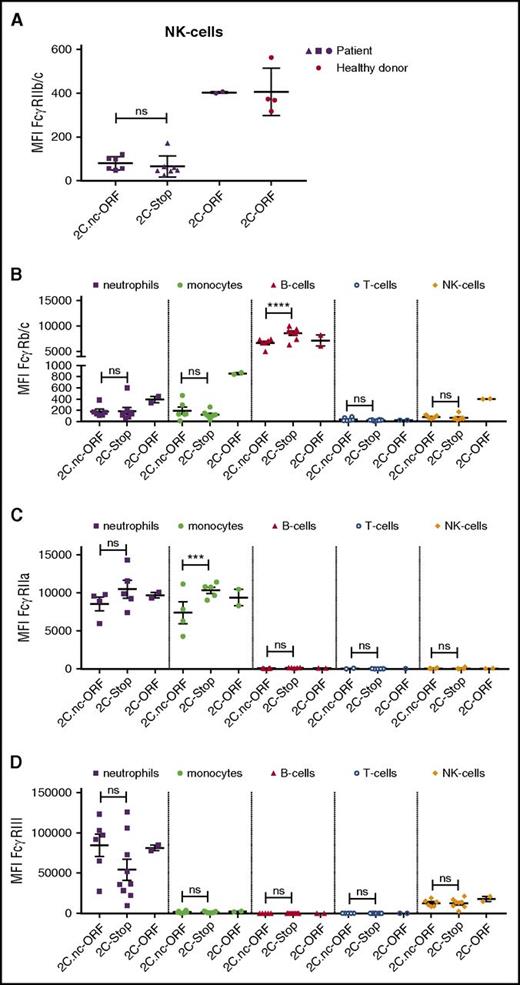
To explain the association between this polymorphism in a nonexpressed gene (Figure 3) and alloimmunization, we assessed FcγR expression profiles of patients with the FCGR2C.nc-ORF polymorphism and compared this to patients with FCGR2C-stop or FCGR2C-ORF by use of flow cytometry (Figure 4; supplemental Table 4). First, we looked at FGCR2C expression. Because the extracellular domain of FcγRIIb and FcγRIIc are identical, we used an antibody that recognizes FcγRIIb/c. We have analyzed natural killer (NK) cells, which do not express FcγRIIb but can express FcγRIIc. In parallel to what can be found in Caucasian donors,50,51 no expression of FcγRIIc was found in patients with FCGR2C-stop or FCGR2C.nc-ORF (Figure 4A).
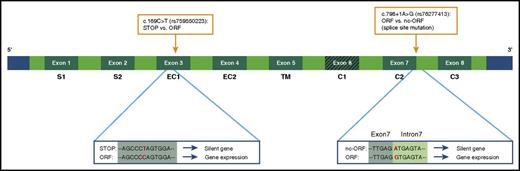
Schematic overview variants in the FCGR2C gene that influence its expression. Because of a premature stop codon, the FCGR2C gene is a nonexpressed pseudogene in FCGR2C-stop individuals. A SNP in exon 3 can lead to an open reading frame (ORF), which results in expression of the gene (FCGR2C-ORF variant). An additional splice site mutation near exon 7 can generate another stop codon, which in turn results in no expression of FcγRIIc.51,72 This last variant is the FCGR2C.nc-ORF polymorphism. The FCGR2C.nc-ORF variant is a combination of the exon 3 SNP and the splice site mutation at exon 7. SNPs are indicated with orange boxes. STOP, stop codon.
Schematic overview variants in the FCGR2C gene that influence its expression. Because of a premature stop codon, the FCGR2C gene is a nonexpressed pseudogene in FCGR2C-stop individuals. A SNP in exon 3 can lead to an open reading frame (ORF), which results in expression of the gene (FCGR2C-ORF variant). An additional splice site mutation near exon 7 can generate another stop codon, which in turn results in no expression of FcγRIIc.51,72 This last variant is the FCGR2C.nc-ORF polymorphism. The FCGR2C.nc-ORF variant is a combination of the exon 3 SNP and the splice site mutation at exon 7. SNPs are indicated with orange boxes. STOP, stop codon.
FcγR expression on various cell types in SCD patients. (A) FcγRIIc expression on natural killer (NK) cells of patients expressing FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 7), and FCGR2C-ORF variant (n = 2). In red, NK cells of healthy controls with the FCGR2C-ORF variant (n = 4). (B) FcγRIIb/c expression on neutrophils, monocytes, B cells, T cells, and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 8), and FCGR2C-ORF variant (n = 2). (C) FcγRIIa expression on neutrophils, monocytes, B cells, T cells and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 4), FCGR2C-stop variant (n = 5), and FCGR2C-ORF variant (n = 2). (D) FcγRIIIa/b expression on neutrophils, monocytes, B cells, T cells, and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 9), and FCGR2C-ORF variant (n = 2). Error bars denote the standard errors of the mean. Stars represent highly significant differences. ***P < .001; ****P < .0001. NK, natural killer; ns, nonsignificant differences.
FcγR expression on various cell types in SCD patients. (A) FcγRIIc expression on natural killer (NK) cells of patients expressing FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 7), and FCGR2C-ORF variant (n = 2). In red, NK cells of healthy controls with the FCGR2C-ORF variant (n = 4). (B) FcγRIIb/c expression on neutrophils, monocytes, B cells, T cells, and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 8), and FCGR2C-ORF variant (n = 2). (C) FcγRIIa expression on neutrophils, monocytes, B cells, T cells and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 4), FCGR2C-stop variant (n = 5), and FCGR2C-ORF variant (n = 2). (D) FcγRIIIa/b expression on neutrophils, monocytes, B cells, T cells, and NK cells on patient cells expressing the FCGR2C.nc-ORF variant (n = 6), FCGR2C-stop variant (n = 9), and FCGR2C-ORF variant (n = 2). Error bars denote the standard errors of the mean. Stars represent highly significant differences. ***P < .001; ****P < .0001. NK, natural killer; ns, nonsignificant differences.
Alternatively, the FCGR2C.nc-ORF polymorphism could be linked to a functional unknown polymorphism elsewhere in the FCGR gene locus. To investigate differential FcγR expression due to a potential unknown polymorphism linked to the FCGR2C.nc-ORF polymorphism, we next assessed FCGR2A, FCGR2/B/C, and FCGR3A/B expression on neutrophils, monocytes, B cells, and T cells among patients with FCGR2C.nc-ORF, FCGR2C.nc-stop, or FCGR2C-ORF. The large spread seen for FcγRIII can be attributed to CNV in tested patients. Interestingly, FcγRIIb/c expression was significantly lower on B cells in FCGR2C.nc-ORF patients than that in FCGR2C.nc-stop patients (Figure 4B). When adding healthy (Caucasian) controls with FCGR2C.nc-ORF or FCGR2C.nc-stop, we still find a lower FcγRIIb/c expression on FCGR2C.nc-ORF individuals (Figure 5A). In addition, FcγRIIa expression was lower on monocytes in FCGR2C.nc-ORF patients than that in FCGR2C.nc-stop patients (Figure 4C). However, when we add healthy (Caucasian) controls with FCGR2C.nc-ORF or FCGR2C.nc-stop, the difference in FcγRIIa expression is no longer present (Figure 5B). Lastly, no significant differences in FcγR expression were found on other cell types (Figure 4A-D).
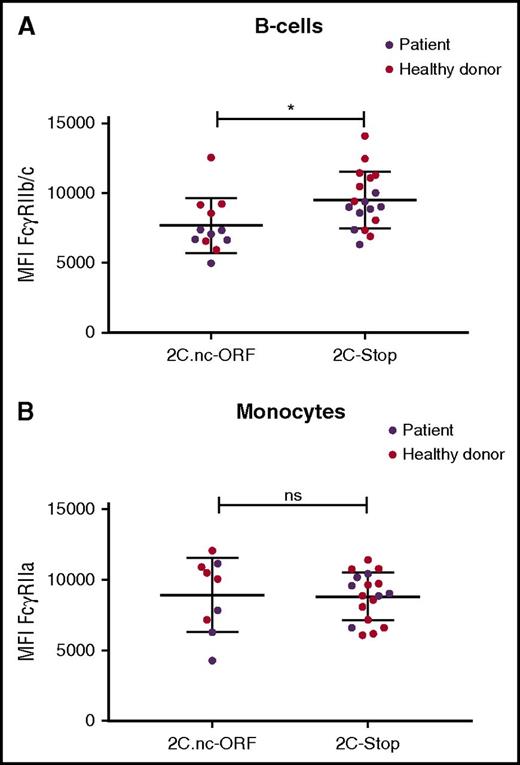
FcγRII expression on B cells and monocytes of SCD patients and healthy controls. (A) FcγRIIb/c expression on B cells on cells expressing the FCGR2C.nc-ORF variant (patients, n = 6; healthy donors, n = 6) or FCGR2C-stop variant (patients, n = 4; healthy donors, n = 10). (B) FcγRIIa expression on monocytes expressing the FCGR2C.nc-ORF variant (patients, n = 4; healthy donors, n = 5) or FCGR2C-stop variant (patients, n = 6; healthy donors, n = 12). *P < .05.
FcγRII expression on B cells and monocytes of SCD patients and healthy controls. (A) FcγRIIb/c expression on B cells on cells expressing the FCGR2C.nc-ORF variant (patients, n = 6; healthy donors, n = 6) or FCGR2C-stop variant (patients, n = 4; healthy donors, n = 10). (B) FcγRIIa expression on monocytes expressing the FCGR2C.nc-ORF variant (patients, n = 4; healthy donors, n = 5) or FCGR2C-stop variant (patients, n = 6; healthy donors, n = 12). *P < .05.
Discussion
This is the first study investigating the association between polymorphisms in the FCGR2/3 gene cluster and alloimmunization against RBC antigens. In a combined cohort of French and Dutch patients with SCD, the FCGR polymorphism FCGR2C.nc-ORF was strongly associated with a lower risk of alloimmunization, conferring a more than 3-fold lower risk for patients with this polymorphism. The protective effect appeared to be strongest when cases with exclusively Rh or K alloantibodies were excluded. These findings suggest that FCGR2C.nc-ORF is involved in the pathophysiology of RBC alloimmunization in SCD, especially for immunization against antigens other than Rh and K.
Current efforts to prevent alloimmunization have been frustrated by the lack of screening methods to prospectively identify patients at high risk of developing alloantibodies. The identification of FCGR2C.nc-ORF as a protective biomarker for alloimmunization may ultimately contribute to the development of appropriate screening tests and targeted prevention of alloimmunization in the near future.
However, our findings do not have implications for alloimmunization against Rh or K antigens. Despite phenotypic matching, this still remains a significant, clinical problem because of genetic variation within RH genes.13,14 Yet when molecular analysis of genetic RH variants becomes more readily available, FCGR2C.nc-ORF may be of great clinical significance as a protective biomarker for alloimmunization beyond Rh or K.
We did not find a significant association between FCGR2/3 polymorphisms and DHTR. It must be noted that DHTR is a multifaceted complication of alloimmunization that is often difficult to diagnose because the symptoms mimic VOC.52 Moreover, the exact mechanism of DHTR is still unclear. It is hypothesized that the pathology is facilitated by FcγR-mediated phagocytosis or ADCC. However, DHTR has also been reported in the absence of detectable alloantibodies.1 Phagocytosis could furthermore be mediated by complement receptors, or hemolysis may be induced via complement activation.53 Therefore, DHTR is a complex process that most likely cannot solely be attributed to FcγRs.
The protective effect of FCGR2C.nc-ORF was strongest when we excluded cases with only Rh or K antibodies. Moreover, in patients with the protective FCGR2C.nc-ORF haplotype that paradoxically did form alloantibodies, the majority primarily formed antibodies of Rh or K specificity, beyond any other RBC antigen-specific antibodies. These data suggest that alloimmunization against the highly immunogenic Rh or K antigens appears to be less dependent on the immunogenetic background of a patient, supporting the clinical relevance of antigen matching for these antigens in patients with SCD. This would also explain why the protective association did not persist in the Dutch subcohort of patients, because the proportion of patients with only Rh or K antibodies was significantly higher here. Alternatively, the lack of association in Dutch patients could also be explained by differences in genetic background due to different ethnic origins of French and Dutch patients with SCD.
The protective marker for alloimmunization that we have identified lies within the FCGR2C gene. FCGR2C expression depends on a combination of 3 minor alleles (Figure 3).51,54 When expressed, FcγRIIc is an activating receptor on NK cells, B cells, monocytes, and neutrophils that can induce innate immune responses such as ADCC.51 Moreover, FcγRIIc counterbalances the inhibitory FcγRIIb on B cells and is thought to enhance antibody responses to immunization,54 which suggests a pivotal role for this gene in (allo)immunization. However, FCGR2C.nc-ORF is a polymorphism in a nonexpressed gene. It is unlikely that this polymorphism is of any direct functional effect. Instead, it may act as a marker for a linked functional variation located elsewhere within the relevant region of chromosome 1. FCGR2C polymorphisms in noncoding variants of the gene have previously been associated with HIV-1 vaccine protection.55 In a follow-up study by Peng et al, it was suggested that these FCGR2C polymorphisms (present in introns) were associated with expression of FcγRIIa and Fc receptor-like A (FCRLA).56 FCRLA is a FCGR homolog that can be selectively expressed on B cells and is suggested to be involved in B-cell development.57
In our phenotype analysis, we found a reduced expression of FcγRIIb on B cells in patients with FCGR2C.nc-ORF. Although this may perhaps somehow provide a clue for a functional explanation of the observed protective effect against RBC alloimmunization, it appears rather counterintuitive in the context of the well-established negative regulatory role of FcγRIIb during BCR signaling in B-cell activation,58 and although alternative functions for FcγRIIb could perhaps also be imagined, such concepts would remain merely speculative. Alternatively, it is possible that FCGR2C.nc-ORF is linked to an unknown polymorphism in the promotor region of FCGR2B, thereby indirectly affecting FcγRIIb expression on B cells. However, additional genotype and phenotype analysis of a larger number of FCGR2C.nc-ORF and FCGR2C.stop patients will be required to further explore this hypothesis.
FCGR2C.nc-ORF and its potential linkage did not seem to induce differential expression of FcγRII or FcγRIII on neutrophils, monocytes, T cells, or NK cells (Figure 3). We have not been able to check FcγR expression on freshly isolated tissue dendritic cells or macrophages, which together with B cells are the principal antigen-presenting cells. Moreover, because the extracellular domain of FcγRIIc is identical to that of FcγRIIb,59 we cannot discriminate between these 2 receptors with flow cytometry using blood cells other than NK cells.47 Although previously reported in systemic lupus erythematosus patients to be relevant on B cells, we have not observed any FcγRIIc expression in these cells either.60 In addition, because of their high degree of homology, our antibodies cannot discriminate between FcγRIIIa and FcγRIIIb (the latter being solely expressed on neutrophils). It is therefore possible that our phenotype analysis was not sensitive enough to identify subtle differences in FcγR expression. Besides inducing differential expression of FcγRs, a polymorphism can also alter the affinity of the receptor for IgG. This type of functional variation will not be detected with our phenotype analysis but requires extensive genotype analysis to find new variants and subsequent affinity studies.
Previous studies have highlighted the ethnic variation in the FCGR2/3 gene locus.54,61-71 Whereas Africa holds the most genetically diverse populations, it is striking that the FCGR2C gene seems to be less polymorphic in our SCD population than in Caucasians.61 FCGR2C is expressed in approximately 18% to 33% of Caucasians,44,61 whereas it is rarely found in African individuals.61 In our cohort, 3% of the patients had the FCGR2C-ORF variant. This frequency may have been too low for us to detect associations for this variant with alloimmunization. It would be interesting to assess this association in a larger SCD cohort, because FCGR2C-ORF has been found to be associated with idiopathic thrombocytopenic purpura, an antibody-mediated autoimmune disease.44 Of note, in comparison with the Caucasian population, we find a high percentage of FCGR2C.nc-ORFs in our cohort. We have previously reported that approximately 20% of individuals with an ORF in exon 3 are FCGR2C.nc-ORF.51 In this cohort, 75.7% of patients with the exon 3 ORF are FCGR2C.nc-ORFs.
We may have missed weak associations of genetic variations that are less common in our study because of our limited sample size. Therefore, this study does not exclude the possibility that other FCGR polymorphisms can be associated with alloimmunization, although they did not reach significance in the current study. Second, our study is specific to alloimmunization in patients with SCD with an originally African background. These results can therefore not directly be extrapolated to other transfused populations and should be the subject of future study. Last, because of the retrospective nature of this study, we cannot fully rule out the possibility that patients have been transfused only at the study institution. Also, patients in the Dutch cohort were exposed to different matching policies over time. Yet the effects on the primary outcomes of this study are estimated to be limited, because this was a case-control study. Patients were selected on the basis of their outcome (alloimmunized or not) and a minimal transfusion exposure for nonalloimmunized controls.
In conclusion, we have found a protective association between FCGR2C.nc-ORF and RBC alloimmunization in SCD. This association was strongest for immunization against antigens other than the highly immunogenic Rh or K. These findings suggest that the FCGR2/3 genes are of less importance in the formation of Rh and K alloantibodies, emphasizing the importance of extended matching for these RBC antigens. Future studies are needed to understand the functional and immunological mechanism behind the protective effect of FCGR2C.nc-ORF on alloimmunization. This may ultimately add to the development of preventive strategies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to acknowledge Lucia Persoon and Alfred van Meurs of the Haga Ziekenhuis; Marjon Cnossen, Sonja Teuben, Cora van Onna-Walraven, and Alice Goncalves da Silva of the Erasmus University Medical Center; and Martijn Veldthuis of Sanquin Blood Supply Foundation for their assistance in the collection and processing of patient blood samples, as well as Philippe Chadebech and Christophe Tournamille for their assistance in the collection of blood samples at the Etablissement Français du Sang (EFS).
S.M.M. was supported by a grant from the Dutch Ministry of Health awarded to T.K.v.d.B. F.P. was supported by grants from the Agence Nationale de la Recherche and from the EFS.
Authorship
Contribution: J.W.R.S. and S.M.M. analyzed the data and wrote the manuscript; J.W.R.S., S.P., A.H., F.P., and K.F. coordinated the collection of DNA samples; S.M.M., S.Q.N. and J.G. performed the MLPA experiments and analyzed the data; S.M.M., J.W.R.S., and M.W.T. performed the statistical analysis; S.M.M. and C.B. performed the phenotype analysis; S.M.M., J.W.R.S., S.Q.N., J.G., B.J.B., K.F., R.v.B., F.P., T.W.K., and T.K.v.d.B. interpreted the data; T.K.v.d.B., T.W.K., F.P., R.v.B., and K.F. supervised the study and edited the manuscript; T.K.v.d.B., F.P., T.W.K., and K.F. designed the study and supervised the interpretation and statistical analysis of the data; and all other authors enrolled patients in the study and revised and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timo K. van den Berg, Department of Blood Cell Research, Sanquin Research, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: t.k.vandenberg@sanquin.nl.
References
Author notes
S.M.M. and J.W.R.S. contributed equally to this work.
F.P. and T.K.v.d.B. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal