Abstract
MYC associated factor X (MAX) is part of the basic-helix-loop-helix (bHLH) leucine zipper transcription factors. MAX forms homo- and hetero-dimers with other bHLH-like factors (e.g. MAD, MXL1 and MYC). The different dimers compete for CACGT DNA E-box consensus sequence (canonical binding site) and other binding sites (non-canonical binding sites) modulating transcriptional activation. The regulation of this process, among others, is based on the different affinity of the formed hetero-dimers to the canonical and non-canonical DNA binding sites and epigenetic modifications in these sequences. In the present study we aim to characterize and understand the role of MAX somatic mutations in the development of Multiple Myeloma (MM).
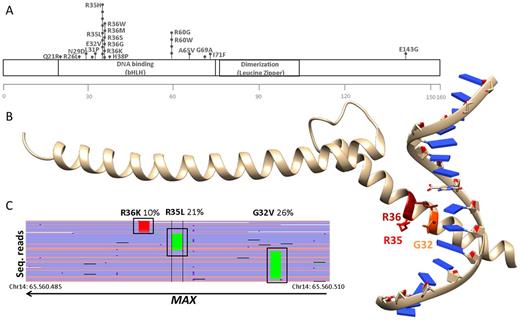
Targeted amplicon sequencing was performed using the M3P (v2.0 or v3.0) gene selection with an average sequencing depth of 700X. We combined data from the different M3P cohorts for a total of 501 M3P patients, 337 newly diagnosed and 164 pre-treated. We also included data from the CoMMpas trial (804 newly diagnosed MM analyzed by WES with 100X average depth). In total 1.305 MM cases were analyzed. The MAX gene was mutated in 33 MM patients (2,5%). Incidence of mutations identified exceeded 5% variant read frequency (VR) and there was no difference in the incidence of the targeted M3P and the WES CoMMpass approach (M3P: 13 of 501 MM cases mutated [2.6%] vs. CoMMpass: 20 of 804 [2.5%]) or between newly diagnosed and pretreated cases (newly diagnosed M3P: 8 of 337 cases [2.4%] vs. pretreated M3P: 5 of 164; [3.0%]). In total, we identified 42 somatic mutations, with 7 patients showing more than one alteration. E.g. in a 73 y/o newly diagnosed MM patient we identified three missense mutations at known MAX hotspot location (p.E32V, p.R35L, p.R36K), all of them in subclonal range with variant reads of 26%, 21% and 10%, respectively. Each mutation exclusively occurred on a different sequencing read and no reads were shared between the mutations. This suggests parallel evolution of different subclones of the disease, and that mutant MAX was virtually present on all tumor cells (confirmed diploid karyotype by cytogenetics). In our cohort we did not observe incidence differences between newly diagnosed and pretreated patients, thus we believe MAX mutations are unlikely to be therapy-related. However, the existence of more than one variant in the gene in 21% of MAX mutated patients (7/33) may indicate a role in disease development. The loss of the protein activity of MAX may provide a survival advantage to the affected tumor clones, possibly due to the binding of other bHLH transcription factors (e.g. MYC) to the E-box, however underlying mechanisms need to be determined. Supporting this hypothesis, 16/42 MAX mutations (38%) affected the integrity of the protein (nonsense, stop gain/loss, tart loss and splice-site donor). The remaining were missense amino-acid changes located in the bHLH DNA binding domaine, including the hotspots R35 (8 mutations), R36 (6 mutations) and R60 (4 mutations), which, according to Wang et al. (Nucleic acid research, 2017) induce a loss of the DNA-binding capacity of the MAX protein. Thus, mutations in MAX in MM patients vastly induce the loss of function or the loss of the protein itself (at least 35/42 (85%)). This inactivation was related with disease development, confirming the role of MAX as a tumor suppressor driver gene in MM.
Figure: A)Location of somatic missense mutations within MAX gene. All except E143G occurred in the DNA binding domaine. Mutations in a.a. G32, R35, R36 and R60 alter DNA binding properties (Wang d et al. Nucleic Acids Research, 2017). B) Three subclonal mutations in a single patient, located at the DNA binding site. The X-ray structure of MAX residues 22-107 bound to DNA (pdb: 5EYO) is shown. C)The mutations belong to different clones. Mutations were detected in independent sequencing reads, covered by the same amplicon, BAM file visualization in IGV sequencing reads aligned to hg19.
Raab: Heidelberg Pharma GMBH: Research Funding. Sonneveld: Celgene Corporation, Amgen, Janssen, Karyopharm, SkylineDx, PharmaMar: Consultancy; Celgene Corporation, Amgen, Janssen, Karyopharm, PharmaMar, SkylineDx: Honoraria; Celgene, Amgen, Janssen, Karyopharm, Takeda: Consultancy, Honoraria, Research Funding. Stewart: Roche: Consultancy; Amgen: Consultancy; Bristol-Myers Squibb: Consultancy; Celgene: Consultancy; Janssen: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal