Key Points
Neutrophils migrate farther and for a longer duration toward a source of P aeruginosa when migrating out of an endothelial lumen.
Blocking IL-6 signaling between endothelial cells and neutrophils decreases neutrophil migration.
Abstract
Neutrophil infiltration into tissues is essential for host defense and pathogen clearance. Although many of the signaling pathways involved in the transendothelial migration of neutrophils are known, the role of the endothelium in regulating neutrophil behavior in response to infection within interstitial tissues remains unclear. Here we developed a microscale 3-dimensional (3D) model that incorporates an endothelial lumen, a 3D extracellular matrix, and an intact bacterial source to model the host microenvironment. Using this system, we show that an endothelial lumen significantly increased neutrophil migration toward a source of Pseudomonas aeruginosa. Surprisingly, we found neutrophils, which were thought to be short-lived cells in vitro, migrate for up to 24 hours in 3D in the presence of an endothelial lumen and bacteria. In addition, we found that endothelial cells secrete inflammatory mediators induced by the presence of P aeruginosa, including granulocyte-macrophage colony-stimulating factor (GM-CSF), a known promoter of neutrophil survival, and interleukin (IL)-6, a proinflammatory cytokine. We found that pretreatment of neutrophils with a blocking antibody against the IL-6 receptor significantly reduced neutrophil migration to P aeruginosa but did not alter neutrophil lifetime, indicating that secreted IL-6 is an important signal between endothelial cells and neutrophils that mediates migration. Taken together, these findings demonstrate an important role for endothelial paracrine signaling in neutrophil migration and survival.
Introduction
Neutrophils act as first responders in an innate immune response, extravasating through the blood vessel endothelium, migrating to sites of infection, performing antimicrobial activities, and recruiting other immune cells. It is known that, after activation, endothelial cells upregulate adhesion molecules1 and secrete cytokines and growth factors2,3 to facilitate neutrophil interaction and transmigration; however, the specific proteins that are secreted after infection are not well characterized, and the effect of these factors on neutrophil migration in interstitial tissues remains unclear.
Neutrophil recruitment is crucial for host defense, but prolonged neutrophil presence can lead to chronic inflammation and tissue damage. It is commonly accepted that neutrophils are short-lived cells with studies reporting in vivo half-lives between 4 and 17 hours.4-9 Recent papers, however, report that neutrophils live longer than initially thought in vivo, with half-lives potentially of 5.4 days.10 This longer lifespan could allow neutrophils to perform more complex processes such as reverse migration-mediated inflammation resolution.11,12 Alternatively, this extended lifetime could be detrimental, leading to tissue damage caused by prolonged systematic inflammation.13 Understanding the mechanisms that modify neutrophil lifetime and motility is, therefore, important for understanding and treating infectious disease and chronic inflammation.4,5,9,14,15
The complex interaction of neutrophils, endothelial cells, and pathogens during an infection could be a target for therapeutics aimed at enhancing or reducing neutrophil survival or migration. Although in vivo infection models intrinsically account for multicellular interactions, their inherent complexities make it difficult to investigate the individual roles of these interactions. Therefore, in vitro models are needed to study neutrophil recruitment to an infection. Microfluidic platforms offer many advantages over traditional in vitro techniques, including the study of primary human cells, direct visualization of cell migration, and inclusion of physiologically relevant structures, such as model blood vessels.16,17 Our group and others have used model endothelial vessels to study neutrophil extravasation across an endothelium to gradients of chemokines.17-21 Although these studies represent a first step in modeling infection, they do not take into account the effect of live bacteria on neutrophils or the endothelium. Importantly, early experiments using endothelial monolayers showed purified lipopolysaccharide did not accurately recapitulate the effect of live bacteria on neutrophil migration or endothelial cell activation,22 indicating that intact bacteria provide a more complex stimulus than purified bacterial products.
We have developed an in vitro model of infection consisting of a model blood vessel, an extracellular matrix, and live bacteria. Using this model, we show that neutrophils extravasating out of an endothelial lumen in the presence of Pseudomonas aeruginosa migrate further and survive longer than neutrophils in a lumen without an endothelium. Furthermore, we show interleukin (IL)-6 is an important promigration signal between endothelial cells and neutrophils in an infection. Our findings demonstrate an important role for neutrophil-endothelial cell interactions in the neutrophil response to infection. Furthermore, this model highlights the importance of studying neutrophil migration in a physiologically relevant environment that integrates multicellular systems and recapitulates in vivo structures.
Methods
LumeNEXT fabrication
The LumeNEXT devices were fabricated as previously described.16 Briefly, the device was formed by patterning 2 polydimethylsiloxane (PDMS) (Dow) layers from SU-8 silicon masters (MicroChem), and bonding them using oxygen plasma (Diener Electronic Femto Plasma Surface System) onto a glass-bottom MatTek dish with a PDMS rod in the chamber (supplemental Methods, available on the Blood Web site).
iEC culture
iCell-endothelial cells (iECs) were purchased from CDI. iECs were maintained in Vasculife Basal maintenance media from LifeLine Cell Technologies supplemented with iCell-Endothelial Cells Medium Supplement (CDI). iECs were plated on cell culture-treated flasks preincubated with 30 µg/mL bovine fibronectin (Sigma-Aldrich). iECs were passaged at 80% confluency and used through passage 5.
Device and collagen preparation
LumeNEXT devices were prepared as previously described.19 Briefly, collagen I (Corning) was neutralized to pH 7.2 (5 mg/mL), pipetted into the devices and around PDMS rods.
After polymerization, the PDMS rods were pulled from the chambers, leaving behind a lumen. Lumens were functionalized with 30 µg/mL bovine fibronectin (Sigma-Aldrich) and seeded with iECs at 2 × 104 cells/µL After 2 hours, unadhered cells were aspirated and replaced with fresh media. Lumens were cultured for 2 days with media exchanges twice daily.
Diffusion experiments
The media in the lumen was exchanged with 1:1 phosphate-buffered saline (PBS):iEC media. P aeruginosa or iEC media was added to the top port of the device and incubated for 2 hours and then set up for imaging. The PBS/iEC media was removed and replaced with a 50 µg/mL solution of 70 kDa fluorescein isothiocyanate (FITC)-Dextran (Sigma-Aldrich).
Neutrophil isolation
All blood samples were drawn according to institutional review board-approved protocols per the Declaration of Helsinki at the University of Wisconsin–Madison. Peripheral neutrophils were isolated from healthy donors, using the MACSxpress Neutrophil Isolation Kit, according to the manufacturer’s directions (Miltenyi Biotec; supplemental Methods). Informed consent was obtained from donors at the time of the blood draw according to our institutional review board. Neutrophils were prestained with calcein AM at 10 nM (Thermo Fisher).
P aeruginosa culture
P aeruginosa PAK and PAK-mCherry strains were used. One colony from an LB plate was grown overnight in 5 mL LB (PAK) or LB-carbenicillin (PAK-mCherry). The next day the culture was diluted 1:5 in fresh media and grown for 1.5 hours. One microliter bacterial culture was pelleted by centrifugation and resuspended in 100 µL iEC media. The optical density (OD) was measured and diluted in iEC media to OD (600 nm), 5. iEC media contain the bacteriostatic antibiotics gentamicin/amphotericin; therefore, in iEC media, the bacteria do not replicate but are alive. The PAK-mCherry strain was used to visualize bacterial diffusion, and the PAK strain was used in migration experiments.
Neutrophil migration
Neutrophils were resuspended at 7.5 × 106 cells/mL in a 1:1 mix of PBS:iEC media and seeded in the lumens. iEC media or P aeruginosa was added to the top port of the device. Lumens were immediately imaged. For blocking antibody experiments, 1.5 × 106 neutrophils were resuspended in 100 µL PBS (Control), 100 µg/mL anti-CD18 (BD Biosciences), 10 µg/mL anti-IL-6Ra (R&D), or 10 µg/mL anti-GM-CSF (R&D). The cells were incubated for 30 minutes at room temperature (anti-CD18)23 or 37°C (anti-IL-6Ra and anti-GMCSF),24 according to published protocols, before dilution with iEC media.
For propidium iodide (PI) experiments, 2 µg/mL PI was pipetted into all 4 device ports and incubated for 1 hours] to ensure diffusion throughout the device. Neutrophils and bacteria in the presence of 2 µg/mL PI were then added as described earlier.
Image acquisition
Migration experiments were imaged on an inverted microscope (Nikon Eclipse TE300), using a Nikon 10×/0.45 (NA) objective, a motorized stage (Ludl Electronica Products), and the acquisition software Metamorph (Molecular Devices). Images were acquired every 10 minutes for 24 hours. PAK-mCherry and FITC-Dextran diffusion experiments were imaged on a Zeiss Z.1 Observer-based inverted spinning disk confocal microscope (Yokogawa) run with Zen software. PAK-mCherry images were collected every 5 minutes for 1 hour, using a Photometrics Evolve EMCCD camera. FITC-dextran diffusion experiments were imaged every minute for 15 minutes, using a Photometrics Coolsnap ES camera for epifluorescence images. All imaging was performed at 37°C.
Image processing and data analysis
Images were transferred to FIJI (ImageJ) for data analysis. Migrated neutrophils were counted using the Analyze Particles function (supplemental Methods). The percentage of PI-positive cells was calculated by counting the number of PI-positive cells in the entire image at each point and dividing by the number of calcein-positive cells at time zero as a loading control. Where large clumps of PI-stained neutrophils made counting difficult, only cells that could be easily identified were counted, leading to an underestimation of the PI staining.
MagPix secretion analysis
Multiplexed protein secretion analysis was performed on lumen-conditioned media from 6 different conditions, using the MagPix Luminex Xmap system (Luminex) with a custom-built bead panel (Thermo Fisher), per the manufacturers’ protocol. Lumens were seeded with endothelial cells, neutrophils, and/or P aeruginosa, according to the conditions, and incubated for 24 hours. For each replicate, media from 6 lumens per condition were collected, pooled, and centrifuged to remove cellular debris. Data are from 4 replicates. Samples were frozen at −20°C from the time of collection until the assay was run. Secreted protein levels were normalized from each replicate with endothelial lumens alone. The heat map was generated using Multi Experiment Viewer software and displays log2 fold change over the condition with endothelial lumens alone.
Statistical analysis
All data represent at least 3 experimental replicates. For migration experiments, the average distance from the lumen and percentage migrated neutrophils were compared at each point, using a Student t test with the Tukey’s HSD (honestly significant difference) post hoc test using SAS software. For the MagPix experiment, levels were compared between conditions using analysis of variance, and the results were summarized in terms of least squared adjusted means and standard errors, using R version 3. P values are labeled as *P < .05, **P < .01, ***P < .001, and ****P < .0001.
Results
Characterization of a 3-dimensional in vitro infection model using an endothelial lumen
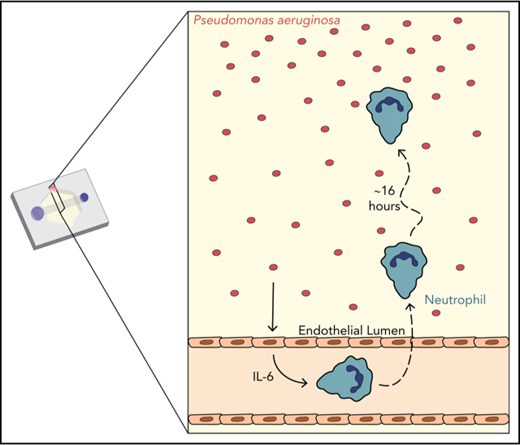
We sought to develop an in vitro infection model that included important features of the infectious microenvironment, including multicellular interactions and physiologically relevant geometries. We employed the recently developed LumeNEXT device16 to generate endothelial lumens, a reductionist model of blood vessels. Neutrophils, seeded inside the lumens, migrate across the endothelium, through a collagen matrix, to the bacteria P aeruginosa (Figure 1A).
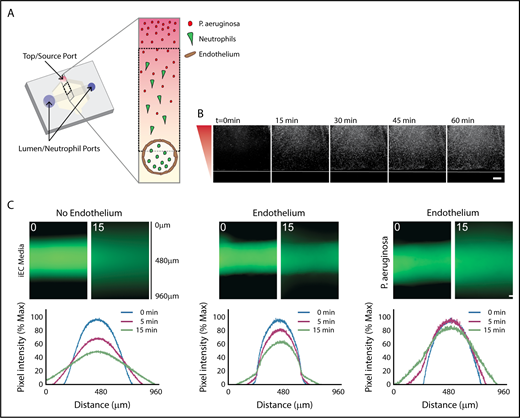
Characterization of in vitro infection model through an endothelial lumen. (A) Schematic of the endothelial lumen infection model shows neutrophils (green) leaving the endothelial lumen and migrating to a source of bacteria (red). Cross-sectional area indicated by the gray box. Black dotted box indicates imaging area. (B) PAK strains expressing mCherry move through the system and reach the lumen by 1 hour. White line indicates the edge of the lumen. Bacterial gradient direction shown in red. (C) FITC-dextran diffusion from lumens at 0 and 15 minutes. Pixel intensity measured across the images and normalized to the intensity of the lumen center at t = 0 minutes. Scale bar, 100 µm for B-C.
Characterization of in vitro infection model through an endothelial lumen. (A) Schematic of the endothelial lumen infection model shows neutrophils (green) leaving the endothelial lumen and migrating to a source of bacteria (red). Cross-sectional area indicated by the gray box. Black dotted box indicates imaging area. (B) PAK strains expressing mCherry move through the system and reach the lumen by 1 hour. White line indicates the edge of the lumen. Bacterial gradient direction shown in red. (C) FITC-dextran diffusion from lumens at 0 and 15 minutes. Pixel intensity measured across the images and normalized to the intensity of the lumen center at t = 0 minutes. Scale bar, 100 µm for B-C.
We first determined how the addition of P aeruginosa altered the established LumeNEXT system.16,19 A strain of P aeruginosa expressing the fluorophore mCherry (PAK-mCherry) was used to visualize bacterial diffusion in the device. We found the bacteria diffused to the endothelial barrier in less than 1 hour, but was excluded from the lumen interior (Figure 1B). Blood vessels have increased permeability during an infection25,26 ; therefore, we wanted to determine barrier function of our endothelial lumens in the presence of P aeruginosa. As previously seen,16,19 lumens without an endothelium had no barrier function, with significant FITC-diffusion seen after 15 minutes, whereas lumens with an endothelium had good barrier function (Figure 1C). Endothelial lumens exposed to P aeruginosa had more FITC diffusion than endothelial lumens with iEC media, but significantly less FITC diffusion than no endothelium, indicating the presence of bacteria increases vessel leakiness, but the endothelium still provides significant barrier function.
P aeruginosa induces neutrophil extravasation and migration
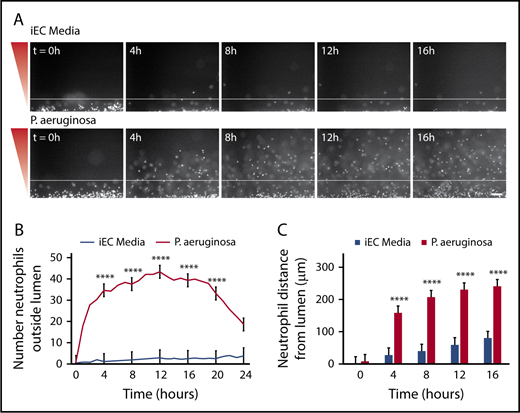
To determine whether P aeruginosa could recruit neutrophils out of the lumen, endothelial lumens were seeded with primary human neutrophils, and P aeruginosa or iEC media were added to the device. We found that neutrophils migrated through the endothelium to P aeruginosa, but not to media alone (Figure 2A; supplemental Video 1). Neutrophils migrated to P aeruginosa throughout the 24-hour imaging period; however, reduced migration was observed after 16 hours. Therefore, we restricted our analysis to the first 16 hours for the remainder of the study. We quantified neutrophil migration in 2 ways. First, the total number of neutrophils outside the lumen, or the number of migrated neutrophils, was counted at 1-hour intervals (Figure 2B). Second, the distance of each neutrophil from the lumen edge, or the neutrophil migration distance, was measured at 4-hour intervals (Figure 2C). In response to P aeruginosa, we found significantly more neutrophils migrated out of the lumen, and that neutrophils migrated significantly farther compared with media alone.
P aeruginosa induces neutrophil extravasation and migration. Neutrophils migrate significantly more to a source of P aeruginosa compared with iEC media alone. (A) Representative images of neutrophils migrating at 4-hour intervals. Neutrophils stained with calcein AM. Gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus standard error of the mean (SEM). (B-C) Data quantified from 9 lumens per condition across 3 independent experiments. Asterisks represents significance between conditions at each point. ****P < .0001.
P aeruginosa induces neutrophil extravasation and migration. Neutrophils migrate significantly more to a source of P aeruginosa compared with iEC media alone. (A) Representative images of neutrophils migrating at 4-hour intervals. Neutrophils stained with calcein AM. Gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus standard error of the mean (SEM). (B-C) Data quantified from 9 lumens per condition across 3 independent experiments. Asterisks represents significance between conditions at each point. ****P < .0001.
Neutrophil extravasation and migration require β2 integrin signaling
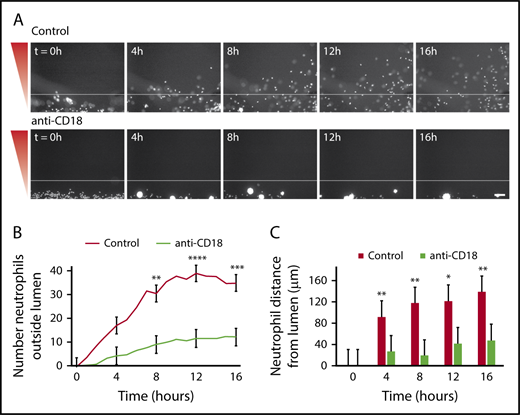
It is known that neutrophil β2-integrins bind to endothelial cell adhesion molecules, and this binding is necessary for neutrophil extravasation.27 To determine whether neutrophils required β2-integrin binding in our endothelial lumens, we tested the effect of a blocking antibody that specifically recognizes the β2-integrin chain (anti-CD18). In the presence of anti-CD18, neutrophils had impaired migration out of the endothelial lumens and aggregated in the intraluminal space (Figure 3A). CD18-blocked neutrophils had a significantly reduced number of migrated neutrophils (Figure 3B) and a significantly reduced neutrophil migration distance (Figure 3C). These findings indicate that β2-integrin-mediated adhesion is crucial for neutrophil extravasation in our model, and active neutrophil-endothelial cell interactions are required for efficient neutrophil migration out of the lumens.
Neutrophil extravasation and migration require β2integrin signaling. Neutrophils were preincubated with PBS (control) or anti-CD18 (β2 integrin) blocking antibody for 30 minutes before starting the migration experiment, and the experiment was run in the continued presence of the antibody. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. Data quantified from 6 lumens (Control) or 5 lumens (anti-CD18) across 3 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
Neutrophil extravasation and migration require β2integrin signaling. Neutrophils were preincubated with PBS (control) or anti-CD18 (β2 integrin) blocking antibody for 30 minutes before starting the migration experiment, and the experiment was run in the continued presence of the antibody. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. Data quantified from 6 lumens (Control) or 5 lumens (anti-CD18) across 3 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
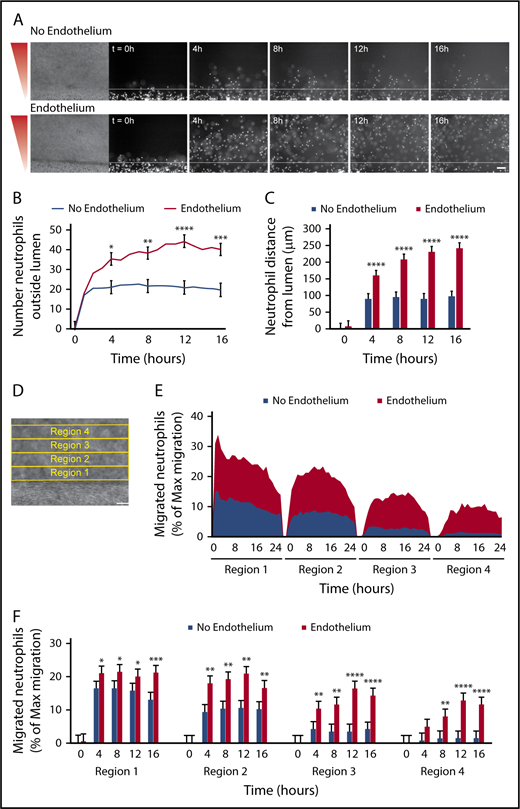
Migration to P aeruginosa requires an endothelium
It is known that inflammatory signals, such as those released from bacteria, activate endothelial cells and alter their secreted protein and adhesion molecule expression.3 Therefore, we hypothesized that endothelial signaling in the presence of bacteria contributes to neutrophil migration. To test this hypothesis, we visualized and quantified neutrophil migration to P aeruginosa out of lumens with and without an endothelium (Figure 4). We found that although some neutrophils migrated out of unlined lumens (lumens without an endothelium), endothelial-lined lumens had vastly more migrated neutrophils (Figure 4A-B). Neutrophils also migrated significantly farther from endothelial lumens compared with unlined lumens (Figure 4C). In addition, time-lapse imaging showed that neutrophils migrate for up to 24 hours after extravasation through the endothelial lumen, whereas neutrophils from an unlined lumen arrest after 4 to 6 hours (supplemental Video 2).
Migration to P aeruginosa requires an endothelium. Neutrophil migration was measured across an unlined lumen (No Endothelium) or an endothelial-lined lumen (Endothelium) to a source of P aeruginosa. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the lumen edge. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. (D-E) The percentage of migrated neutrophils relative to the maximum number of migrated neutrophils at any point per condition was determined in four, 100-µm regions at increasing distance from the lumen (D) and plotted as an area graph (E). (F) Statistical significance for migrated neutrophils (% maximum migration) in each region was determined at 4-hour intervals. Bars represent the mean plus SEM. All data were quantified from 13 lumens (No Endothelium) or 14 lumens (Endothelium) across 5 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
Migration to P aeruginosa requires an endothelium. Neutrophil migration was measured across an unlined lumen (No Endothelium) or an endothelial-lined lumen (Endothelium) to a source of P aeruginosa. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the lumen edge. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. (D-E) The percentage of migrated neutrophils relative to the maximum number of migrated neutrophils at any point per condition was determined in four, 100-µm regions at increasing distance from the lumen (D) and plotted as an area graph (E). (F) Statistical significance for migrated neutrophils (% maximum migration) in each region was determined at 4-hour intervals. Bars represent the mean plus SEM. All data were quantified from 13 lumens (No Endothelium) or 14 lumens (Endothelium) across 5 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
We next sought to quantify neutrophil migration by combining the number of neutrophils migrating and the migration distance into 1 measurement. To do this, we determined the number of migrated neutrophils in four, 100-µm regions of increasing distance from the lumen (Figure 4D). This analysis showed at least 10% of migrating neutrophils from an endothelial lumen reach region 4 by the end of the experiment, whereas only about 1% of migrating neutrophils from an empty lumen reach this region (Figure 4E). This quantification also shows the movement of neutrophils through regions 1 to 4; as the percentage of neutrophils in regions 3 and 4 increases over time, the percentage of neutrophils in region 1 decreases. We performed statistical analysis at 4-hour intervals and found neutrophil migration with an endothelial lumen was significantly higher in the first 3 regions after 4 hours and in the fourth region after 8 hours (Figure 4F). Together, these results show an endothelial lumen is critical for efficient and persistent neutrophil migration to P aeruginosa and indicate that neutrophil-endothelial interactions promote neutrophil interstitial migration in an infection.
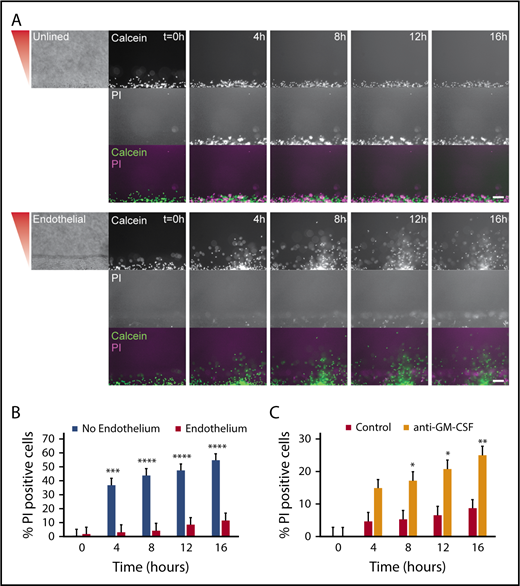
An endothelial lumen increases neutrophil lifetime
On the basis of our finding that an endothelium caused neutrophils to migrate for significantly longer durations in the presence of P aeruginosa, we hypothesized that endothelial cells were increasing neutrophil survival. To quantify neutrophil lifetime in the presence of endothelial cells and P aeruginosa, we imaged neutrophil migration out of unlined and endothelial-lined lumens in the presence of the live/dead cell stain PI. We found neutrophils from endothelial lumens had almost no PI staining during the 16-hour experiment, indicating these neutrophils lived much longer than neutrophils from unlined lumens, which became PI positive at 4 hours. (Figure 5A). The images showed PI-positive endothelial cells in the endothelial lumen, but no PI-positive neutrophils in the periphery. The percentage of PI-positive cells clearly shows neutrophils in unlined lumens have significantly more death after 4 hours (Figure 5B). GM-CSF is known to increase neutrophil lifetime in vitro28-30 ; therefore, we tested its role in promoting neutrophil lifetime. We found that the percentage of PI-positive neutrophils in lumens incubated with an anti-GM-CSF blocking antibody was significantly higher than in uninhibited lumens (Figure 5C). Interestingly, there was no migration defect seen after inhibition with anti-GM-CSF (supplemental Figure 1). These results indicate that in addition to increasing neutrophil migration, endothelial cell interactions increase neutrophil lifetime in the presence of P aeruginosa, likely through secretion of GM-CSF.
An endothelial lumen increases neutrophil lifetime. Propidium iodide staining was used to monitor neutrophil lifetime in unlined and endothelial-lined lumens. (A) Calcein AM was used to stain neutrophils (green), and PI was used to stain dead cells (magenta). Phase images (left) are shown for lumen edge visualization. The gradient direction is indicated in red. Images are representative of 4 independent experiments. Scale bar, 100 µm. (B) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. (C) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. Bars represent mean plus SEM. All data were quantified from 12 lumens (No Endothelium) or 11 lumens (Endothelium) across 4 independent experiments, and from 9 lumens (Control) or 8 lumens (anti-GM-CSF) across 3 independent experiments. Asterisks represent significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
An endothelial lumen increases neutrophil lifetime. Propidium iodide staining was used to monitor neutrophil lifetime in unlined and endothelial-lined lumens. (A) Calcein AM was used to stain neutrophils (green), and PI was used to stain dead cells (magenta). Phase images (left) are shown for lumen edge visualization. The gradient direction is indicated in red. Images are representative of 4 independent experiments. Scale bar, 100 µm. (B) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. (C) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. Bars represent mean plus SEM. All data were quantified from 12 lumens (No Endothelium) or 11 lumens (Endothelium) across 4 independent experiments, and from 9 lumens (Control) or 8 lumens (anti-GM-CSF) across 3 independent experiments. Asterisks represent significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
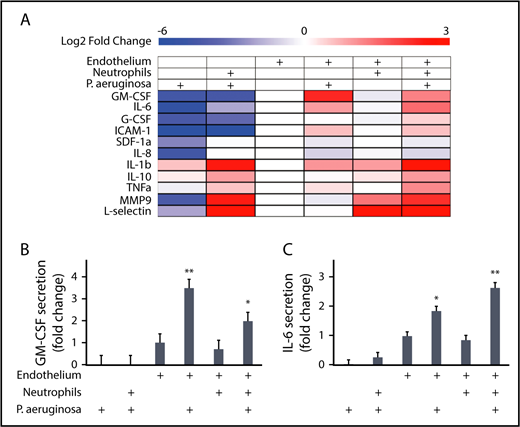
Endothelial cells secrete IL-6 in response to Pseudomonas
We were next interested in determining what factors endothelial lumens produce in response to P aeruginosa that promote neutrophil migration. We used a multiplexed ELISA (MagPix) panel to compare the levels of immunologically relevant proteins in lumen-conditioned media (Figure 6A). The total protein levels measured in each condition represent the sum of protein secretion and protein consumption; although these 2 processes cannot be separated in the analysis, we can still gain insight into secretion patterns by comparing between conditions (Figure 6A).
Endothelial cells secrete IL-6 and GM-CSF in response to P aeruginosa. A multiplexed enzyme-linked immunosorbent assay screen was run on lumen-conditioned media from 6 conditions with different combinations of neutrophils, P aeruginosa, and an endothelium. (A) The log2 fold change of protein levels over the endothelial lumen-only condition are displayed as a heat map. Blue indicates lower expression and red indicates increased expression. Condition combinations are indicated on the top of the heat map. Factors measured are listed on the left of the heat map. Four factors (VEGFD, VEGFA, P-selectin, and RANKL) were below the level of detection and are not displayed. (B) The levels of GM-CSF expressed as a fold change over the condition with only an endothelial lumen were plotted. Bars represent least-squared mean plus SEM. Condition combinations are indicated below the graph. (C) The levels of IL-6 expressed as a fold change over the condition with only an endothelial lumen were plotted. Bars represent least-squared mean plus SEM. Condition combinations are indicated below the graph. Data in B and C are the same as for these proteins in A, but displayed with statistics. All data represent 4 independent replicates, with media from 6 lumens pooled per replicate. Asterisks show significance relative to all other conditions, with the level of significance represented as the lowest level for any condition. *P < .05 and **P < .01.
Endothelial cells secrete IL-6 and GM-CSF in response to P aeruginosa. A multiplexed enzyme-linked immunosorbent assay screen was run on lumen-conditioned media from 6 conditions with different combinations of neutrophils, P aeruginosa, and an endothelium. (A) The log2 fold change of protein levels over the endothelial lumen-only condition are displayed as a heat map. Blue indicates lower expression and red indicates increased expression. Condition combinations are indicated on the top of the heat map. Factors measured are listed on the left of the heat map. Four factors (VEGFD, VEGFA, P-selectin, and RANKL) were below the level of detection and are not displayed. (B) The levels of GM-CSF expressed as a fold change over the condition with only an endothelial lumen were plotted. Bars represent least-squared mean plus SEM. Condition combinations are indicated below the graph. (C) The levels of IL-6 expressed as a fold change over the condition with only an endothelial lumen were plotted. Bars represent least-squared mean plus SEM. Condition combinations are indicated below the graph. Data in B and C are the same as for these proteins in A, but displayed with statistics. All data represent 4 independent replicates, with media from 6 lumens pooled per replicate. Asterisks show significance relative to all other conditions, with the level of significance represented as the lowest level for any condition. *P < .05 and **P < .01.
Our analysis revealed groups of proteins with similar secretion patterns. For example, l-selectin and MMP9 had increased levels in conditions with neutrophils, whereas G-CSF and ICAM-1 showed increased levels in the presence of an endothelium. We also found that neutrophils secrete high levels of IL-1β only in the presence of P aeruginosa. Most relevant in determining what factors endothelial cells secrete that might be promoting neutrophil functions during infection, GM-CSF and IL-6 levels were increased only in the combined presence of endothelial lumens and P aeruginosa. Although GM-CSF (Figure 6B) and IL-6 (Figure 6C) were both present at significantly higher levels in endothelial lumens with P aeruginosa compared with all other conditions, IL-6 levels were further increased in the presence of neutrophils (Figure 6C). This indicates that although IL-6 is secreted by endothelial cells alone in the presence of P aeruginosa, signaling between the endothelial cells and neutrophils leads to a further increase in IL-6 secretion.
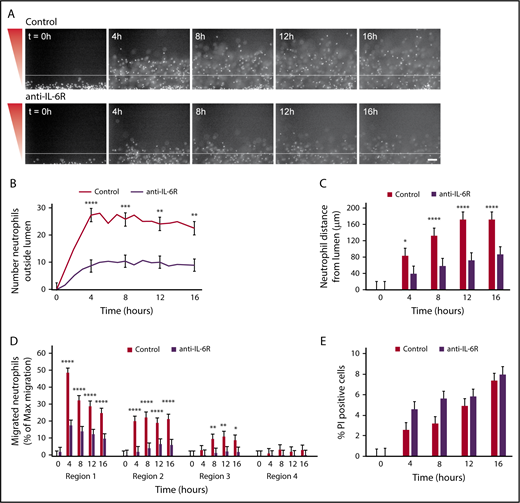
Neutrophil migration requires IL-6 signaling
Given our migration data showing increased neutrophil migration out of an endothelial lumen toward P aeruginosa and our MagPix data showing endothelial lumens produce IL-6 in response to P aeruginosa, we hypothesized that neutrophil migration outside the endothelial lumen requires IL-6 signaling. To test this hypothesis, we used a blocking antibody against the IL-6 receptor (anti-IL-6R) and found neutrophils migrated significantly less efficiently after incubation with anti-IL-6R compared with control neutrophils (Figure 7A; supplemental Video 3). There were significantly fewer neutrophils outside the lumen with IL-6R inhibition (Figure 7B). Furthermore, antibody-blocked neutrophils had a significantly reduced migration distance at all points compared with unblocked neutrophils (Figure 7C). This pattern of migration resembled neutrophil migration out of an unlined lumen. Therefore, we quantified neutrophil migration, using our combined approach, and looked at migration within the 4 regions previously described (Figure 4D). Again, we found significantly reduced migration for neutrophils incubated with anti-IL-6R compared with uninhibited neutrophils (Figure 7D). Finally, we imaged uninhibited and antibody-blocked neutrophils in the presence of PI to determine whether IL-6 signaling altered neutrophil lifetime, and found no significant difference in the percentage of PI-positive cells between cells preincubated with anti-IL-6R and uninhibited neutrophils (Figure 7E). Altogether, our data demonstrate that IL-6 is a crucial signaling molecule between endothelial cells and neutrophils that promotes neutrophil migration out of an endothelial lumen to P aeruginosa.
Neutrophil migration requires IL-6 signaling. Neutrophils were preincubated with PBS (control) or anti-IL-6R blocking antibody for 30 minutes before starting the migration experiment, and the experiment was run in the continued presence of the antibody. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. (D) The number of migrated neutrophils (% maximum migration) in each region was determined at 4-hour intervals. (E) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. Bars represent the mean plus SEM. Data quantified from 11 lumens (Control) or 14 lumens (anti-IL-6R) across 4 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
Neutrophil migration requires IL-6 signaling. Neutrophils were preincubated with PBS (control) or anti-IL-6R blocking antibody for 30 minutes before starting the migration experiment, and the experiment was run in the continued presence of the antibody. (A) Representative images of neutrophil migration. Neutrophils stained with calcein AM. Bacterial gradient direction shown in red. White line indicates the edge of the endothelial lumen. Scale bar, 100 µm. (B) The number of neutrophils outside the lumen was counted at 1-hour intervals, using particle analysis in FIJI. (C) The distance from the lumen edge was measured for all in-focus neutrophils outside the lumen at 4-hour intervals. Each bar represents the mean plus SEM. (D) The number of migrated neutrophils (% maximum migration) in each region was determined at 4-hour intervals. (E) The number of PI-positive cells in the whole image was counted using FIJI and normalized to the number of calcein AM-positive cells at t = 0 hours. Bars represent the mean plus SEM. Data quantified from 11 lumens (Control) or 14 lumens (anti-IL-6R) across 4 independent experiments. Asterisks represents significance between conditions at each point. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
Discussion
In this study, we developed an in vitro microfluidic system for studying neutrophil migration that recapitulates many important aspects of an in vivo infection by studying neutrophils in a model endothelial blood vessel with a source of intact bacteria. Using this system, we found that neutrophil migration and survival are greatly increased by endothelial cell signaling in response to P aeruginosa. In particular, we found endothelial cells produce GM-CSF and IL-6 in response to P aeruginosa and that these signals specifically increase neutrophil lifetime and migration, respectively.
It is known that interactions between neutrophils and the vascular endothelium are important for neutrophil recruitment to an infection.27 The inclusion of a model endothelial blood vessel in our system allowed us to directly study this interaction. The use of an endothelial lumen as opposed to an endothelial monolayer was important, as it has been shown that culture geometry alters the secretion profile of endothelial cells.31 This was especially important in our system, as differences in the level of specific secreted proteins, such as IL-6, increased neutrophil migration. It was previously shown that integrin-mediated neutrophil-endothelial cell binding leads to outside-in signaling that promotes neutrophil migration,32,33 and we have shown that β2 integrin binding is necessary for neutrophil migration in our system. Furthermore, it is known that transendothelial migration alters the surface receptors of neutrophils,33,34 which could increase their ability to migrate through extracellular matrix. Therefore, further studies are needed to determine whether the presence of endothelial-secreted proteins such as IL-6 alone are sufficient to increase neutrophil migration or whether transendothelial migration is also necessary.
Extended neutrophil lifespans are required for the recently recognized neutrophil reverse migration mechanisms involved in inflammation resolution. It was previously thought that after an immune response, neutrophils died and were cleared at the site of infection. However, it was shown in both zebrafish and murine models that after an inflammatory event, neutrophil infiltration is also resolved through reverse migration away from the site of inflammation.11,12,35 It was further shown that reverse migrated neutrophils go to secondary sites of inflammation36 and to the bone marrow.12,13 This is clinically relevant, as reverse transmigrated neutrophils were significantly more prevalent in patients with acute pancreatitis-associated lung injury, illustrating the potential for tissue damage caused by long-lived neutrophils.13 These processes require long-lived neutrophils in vivo, and our data suggest this increased lifetime may be partly a result of interactions with the vascular endothelium.
Although it is known that GM-CSF prolongs neutrophil survival in vitro by delaying apoptosis,28-30 the role of IL-6 in promoting neutrophil migration is less clear. It is known that endothelial cells in culture express IL-6 in response to activating signals such as proinflammatory molecules2,3,37-39 and infection40 , but the reported effects of IL-6 on neutrophil survival in vitro are inconsistent with papers indicating IL-6 has no effect,28 increases,41,42 or decreases43 neutrophil lifetime. Our results suggest that during an infection, IL-6 secretion by endothelial cells enhances neutrophil migration, but not survival, in response to infection.
Interestingly, a study using mice infected with the H1N1 influenza virus showed increased IL-6 levels played a protective role by promoting neutrophil survival and enhancing neutrophil-mediated virus clearance.44 Our data suggest this increase in IL-6 could be in part a result of secretion by endothelial cells. This could have important clinical implications, as inhibitors of IL-6 are currently being used to treat several types of cancer45-47 and rheumatoid arthritis.48,49 Although these inhibitors have been successful in treating disease, they are also associated with an increased incidence of serious infection.50 As we have shown, IL-6 secretion is critical for efficient neutrophil migration in our system, which could explain the increased rates of bacterial infection seen in patients treated with IL-6 inhibitors.
Secretion of IL-6 and GM-CSF was dependent on endothelial cell activation by P aeruginosa. Much of the current in vitro research on neutrophil-endothelial cell interaction relies on the use of purified molecules to model inflammation.2,3 Pro-inflammatory molecules such as tumor necrosis factor α or bacterial products such as lipopolysaccharide are used to activate endothelial cells,2,3 and chemoattractants such as fMLP or IL-8 are used to induce neutrophil migration.17,19,20 Although these methods have revealed important neutrophil-endothelial interactions, they do not accurately recapitulate the complex signals induced by pathogens. This fact is highlighted by studies showing differences in neutrophil recruitment and endothelial cell signaling in the presence of intact bacteria vs bacterial products.22 We found, in our system, that a source of P aeruginosa resulted in significantly more robust neutrophil migration compared with our previous studies using fMLP or IL-8 as a chemoattractant.19 Although we do not know exactly what signals P aeruginosa is secreting in our system, it was shown in a mouse model that P aeruginosa can cause a release of eicosanoids from endothelial cells, which lead to neutrophil-mediated inflammation.26 In addition, it has been shown that outer membrane vesicles secreted by Escherichia coli, another gram-negative bacteria, cause increased expression of several molecules, including IL-6, in endothelial cells.51,52 It is possible the P aeruginosa is having a similar effect in our model, as the expression of IL-6 by endothelial cells was only elevated in the presence of bacteria. Although the exact mechanisms of neutrophil and endothelial cell activation by P aeruginosa are not known, it is clear that inclusion of pathogen rather than inflammatory molecules is important for studying neutrophil-endothelial interactions during infection.
Future work is needed to determine whether neutrophil-endothelial cell interactions promote neutrophil migration in other infections. The importance of endothelial cell signaling on neutrophil migration in disease is also a key avenue of future study. Moreover, the device could be a useful tool for investigating neutrophil migration to infection with neutrophils from patients suffering from chronic inflammation or infections. Furthermore, although commercially available induced pluripotent stem cell–derived endothelial cells were used in this study, in the future, patient-derived induced pluripotent stem cell–derived endothelial cells could be used to study neutrophil-endothelial cell interactions and neutrophil migration in a disease model. In the future, models of infection that accurately represent the in vivo microenvironment while allowing for the analysis of specific interactions, such as the system developed in this study, will allow us to determine the molecular mediators of neutrophil migration and the role of the endothelium and bacteria in this process.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health, National Cancer Institute through R01 CA186134, the National Institutes of Health, National Institute of General Medical Sciences R35 GM1 18027 01, in addition to National Institutes of Health, National Heart, Lung, and Blood Institute Hematology T32 HL07899 at the University of Wisconsin–Madison.
Authorship
Contribution: L.E.H., P.N.I., D.J.B., and A.H. designed the research and wrote the manuscript; L.E.H. and P.N.I. performed the experiments; and L.E.H., P.N.I., D.J.B., and A.H. analyzed and interpreted the data.
Conflict-of-interest disclosure: D.J.B. is a board member and stockowner of Tasso, Inc., and a stockowner of Bellbrook Labs, LLC; a founder, stockowner, and consultant of Salus Discovery LLC; and an advisor and stockowner of Lynx Biosciences, LLC. D.J.B. also holds equity in Bellbrook Labs, LLC, Tasso, Inc., Salus Discovery, LLC, Stacks to the Future, LLC, and Onexio Biosystems LLC. The remaining authors declare no competing financial interests.
Correspondence: Anna Huttenlocher, University of Wisconsin–Madison, 1550 Linden Dr, Room 4225, Madison, WI 53706; e-mail: huttenlocher@wisc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal