In this issue of Blood, Roberts et al describe a preclinical mouse model of acute lymphoblastic leukemia (ALL) caused by the ETV6-NTRK3 fusion gene.1
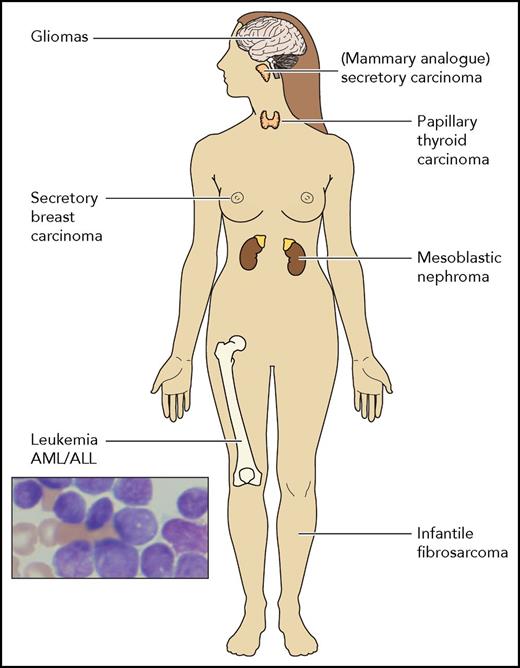
A schematic presentation of the different types of cancers reported to harbor the ETV6-NTRK fusion gene. Other NTRK fusion translocations have been rarely reported in many diverse cancers including soft tissue sarcomas, lung colon and pancreatic cancers, cholangiocarcinomas, and more. AML, acute myeloid leukemia.
A schematic presentation of the different types of cancers reported to harbor the ETV6-NTRK fusion gene. Other NTRK fusion translocations have been rarely reported in many diverse cancers including soft tissue sarcomas, lung colon and pancreatic cancers, cholangiocarcinomas, and more. AML, acute myeloid leukemia.
The mouse leukemias were highly sensitive to the specific tropomyosin receptor kinase (TRK) inhibitor that has recently demonstrated significant clinical efficacy in treatment of pediatric and adult solid tumors with NTRK fusions.2,3
Philadelphia chromosome (Ph)–like ALLs are high-risk leukemias characterized by a similar gene expression to that of BCR-ABL1–positive ALLs.4-6 They are caused by somatic genomic events activating the JAK-STAT or ABL signaling pathways. Clinical trials testing the efficacy of adding JAK or ABL tyrosine kinase inhibitors to chemotherapy are ongoing (eg, www.clinicaltrials.gov, #NCT01164163). Among the rearrangements identified in Ph-like ALLs, isolated cases with ETV6-NTRK3 have been reported.4,7
Originally discovered in infantile fibrosarcoma,8 the ETV6-NTRK3 fusion gene is detected in multiple types of tumors (see figure). The NTRK1-3 genes encode the TRK family of tyrosine kinase receptors that are important for neural development. Other fusion translocations involving any of the TRK receptors are also rarely detected in many solid tumors and cause constitutive activation of the kinase domain and oncogenic transformation. Recent RNA sequencing uncovered NTRK fusions in 8 of 7311 hematological malignancies including acute myeloid and lymphoid leukemias, histiocytic and dendritic cell neoplasms, and multiple myeloma.7 Thus, oncogenic NTRK fusions are identified in many types of cancers.
The traditional classification of tumors is based on histology and tissue of origin. Because of the remarkable progress in genomic characterization of tumors, a complementary molecular classification of cancer based on causative somatic genetic events has been suggested. Precision medicine drugs target the products of these genetic events regardless of tumor histology. A novel specific inhibitor of all TRK proteins, larotrectinib, has recently shown durable responses in the majority of pediatric and adult patients with TRK fusion positive solid tumors of diverse types.2,3 Treatment with larotrectinib has had a reasonable safety profile. Notably, there were no responses in TRK-negative tumors. These dramatic results in phase 1/2 clinical trials demonstrate the premise of genomic-based precision medicine.
These discoveries raise diagnostic and therapeutic challenges that are fundamental to the new era of precision genomic medicine. Which neoplasms should be analyzed for TRK fusions and what is the preferred methodology? How should the extremely rare patient with a hematological malignancy with TRK fusion be treated, as the above-mentioned studies included patients with solid tumors only? Because of the rarity of suitable patients, a clinical trial for acute leukemias with TRK fusions is very unlikely.
The preclinical model reported in this issue of Blood demonstrates significant activity of TRK inhibitors against a patient-derived xenograft of ETV6-NTRK3 ALL. In a recent case report of a patient with secondary acute myeloid leukemia with the ETV6-NTRK2 fusion, elimination of the ETV6-NTRK2 clone was observed after 10 weeks of therapy with oral larotrectinib.7 However, the patient relapsed with leukemic cells carrying other oncogenic proteins, demonstrating the problem of clonal heterogeneity. We believe that based on these preclinical and clinical trials with TRK inhibitors, and on the paradigm of BCR-ABL1–positive ALL,9 it is reasonable to perform a “single patient clinical trial” with TRK inhibitors for chemotherapy-resistant NTRK fusion–positive hematological malignancy.
Conflict-of-interest disclosure: The authors declare no competing financial interests.