In this issue of Blood, Huet et al analyze the real-life applicability of “knowledge banks“ (see figure); large databases of patients with acute myeloid leukemia (AML) treated with high-dose chemotherapy in a tertiary care center, containing genomic data matched to clinical variables, treatments, and outcomes.1,2
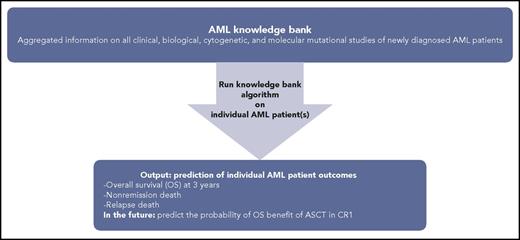
Knowledge bank approach to individualizing AML patient therapy. Schematic representing use of an AML knowledge bank to predict an individual AML patient’s journey.
Knowledge bank approach to individualizing AML patient therapy. Schematic representing use of an AML knowledge bank to predict an individual AML patient’s journey.
Patients with AML usually present acutely, often with life-threatening symptoms requiring prompt diagnosis and initiation of therapy. Although studies have conflicting results on the effect of waiting for complete genetic testing results, the acuity of the disease usually limits the ability to wait for the results before therapy is initiated because of an average turnaround time of approximately 10 to 21 days for cytogenetic and molecular mutational testing results.3,4 It is clear that there are multiple distinct clinicogenomic categories of AML, as evidenced by the various classifications, including the 2016 World Health Organization classification of myeloid neoplasms and acute leukemia.5 Papaemmanuil et al analyzed the impact of multiple genomic mutations and their interactions on prognosis in AML.6 Their study proposed a novel genomic classification of 11 different categories of AML, each with diagnostic features and differing clinical outcomes based on these results.
Another dilemma that confronts oncologists is when an AML patient should receive an allogeneic stem cell transplant (ASCT). The maximum benefit of an ASCT is usually in first complete remission (CR1), but this approach has to be weighed against the treatment-related mortality and chronic morbidity, usually from graft-versus-host disease. This is further complicated by the fact that some patients can be salvaged after first relapse and offered the benefit of ASCT at that time.7,8
These important clinical decision-making conundrums were elegantly approached by Gerstung et al in about 1500 AML patients from 3 randomized clinical trials with validation using The Cancer Genome Atlas cohort.2 They combined driver point mutations and data from the clinical trials database to generate a comprehensive knowledge bank, tested an algorithm to understand the utility of the knowledge bank for generating predictions tailored to the individual patient, and then compared the prediction to the actual outcomes. They also demonstrated how the knowledge bank approach could estimate the likelihood of various clinical end points under different treatment strategies.
The main outcome Gerstung et al looked at when using their algorithm was overall survival (OS). They focused specifically on the benefit (or lack thereof) of ASCT in CR1 and its effect on OS. They found that, when their algorithm is applied to individual patients as opposed to the whole population, it can improve clinical decision-making for that patient. About one-third of AML patients in the study would have had a change in their treatment plan compared with what would be recommended using the European Leukemia Net (ELN) classification. This included significant optimization of the use of ASCT in CR1 for maximum OS benefit. The model also offered improved granularity into different patient outcomes during AML treatment, including death without complete remission, nonrelapse death (mostly treatment related), and death after relapse, as well as survival in induction, CR1, and after relapse.
How does the work of Gerstung et al affect clinical management of AML patients not enrolled in clinical trials ? Huet et al use a similar approach, but rather than using an ideal cohort as represented by clinical trial and The Cancer Genome Atlas populations, they looked at 155 real-world AML patients that received induction chemotherapy with anthracycline and cytarabine, with a proportion of these patients (n = 70) receiving ASCT as part of their AML therapy. They used a similar “knowledge bank” approach, but as expected in the real world, had to take into account missing clinical, cytogenetic, and genomic data. Their intent was to see how well this approach would work in the real-world setting. They assessed the potential of the knowledge bank approach by using an algorithm (Huet et al, supplemental Methods1 ) to predict overall survival at 3 years and compared it with the observed outcome. They found that the knowledge bank approach was better at predicting patient outcomes as compared with the ELN 2010 or 2017.7,9 They also looked at the knowledge bank’s ability to predict outcomes such as nonremission death, relapse death, and nonrelapse death and found that it was able to do in the case of relapse death and nonremission death. The question of the benefit of ASCT could also not be addressed in this study because of small sample size. The lack of missing molecular data (20%) did not seem to have a significant effect on the knowledge bank algorithm, likely because critical genetic mutations that contribute to OS in AML were comprehensively captured and the algorithm was able to compensate for certain missing data.
The paper by Huet et al, describing the ability to use the knowledge bank approach in which we can aggregate clinical and genomic data to make more precise individualized predictions about an AML patient’s outcome with various therapies, represents the ultimate culmination of precision and evidence-based medicine. The refinement of the use of ASCT, which was not demonstrated by this particular paper but can presumably be done using larger sample sizes, will have an enormous effect on AML therapy and on optimizing the use of our available treatment resources as we make incremental steps forward in fighting this disease.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal