Acute lymphoblastic leukemia (ALL) is a prevalent hematopoietic malignancy that requires urgent medical intervention to prevent inferior outcome. ALL is characterized by recurrent structural rearrangements, whole chromosome copy number changes and microdeletions of prognostic value. Currently an extensive panel of tests is required, including karyotype, FISH, whole genome arrays, Multiple Ligation-dependent Probe Amplification (MLPA) and PCR-based diagnostic methods. Testing is thus costly, laborious and time-consuming and due to the large number of tests required cascade testing is often performed resulting in lengthy turn-around-times (TAT). This does not conform to treatment protocols that require rapid results to stratify patients into different treatment arms and to identify those who may benefit from "targeted" therapies, such as the Ph-like ALLs. It is therefore interesting to examine new technologies which can simultaneously detect numerical and structural abnormalities in one assay. Such approaches have not yet been extensively introduced into routine diagnostics but implementation would not only be beneficial in reducing TAT but would simplify the analytical pathways and theoretically increase the diagnostic yield.
One such novel technology is genome imaging of ultra-long linear molecules, enzymatically labeled at specific sequence motifs. The Saphyr genome imaging device (Bionano Genomics) detects numerical and structural variants (>500bp) with a TAT of less than one week. The aim of the current study is to assess whether implementation of this technology could (partially) replace current analytical strategies.
Ten diagnostic T- and 10 diagnostic B-ALL cases were included in this series. All samples were analyzed with the Rare Variant Pipeline (RVP) and filters were set to detect chromosomal aberrations of greater than 5 Mb. For deletions, the limit of detection was set at 500 bp but only regions encompassing clinically relevant loci were investigated.
There was an overall excellent concordance between the results:
Bionano results were concordant with MLPA, a technique routinely used for rapid detection of key CNAs.
All but one recurrent translocations identified by routine strategies were correctly identified by Bionano which failed to detect a STIL-TAL1 rearrangement in one of the two cases with this abnormality.
Bionano further identified:
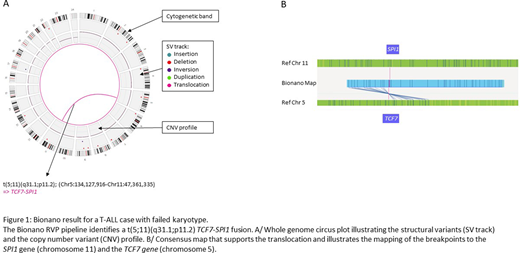
A t(5;11) TCF7-SPI1 in a case with a failed karyotype (subsequently confirmed by another method) (Figure 1).
A t(12;22) ZNF384-EP300 fusion in a case with a normal karyotype. This recurrent rare fusion was confirmed by FISH.
In general there was an excellent concordance of whole/partial chromosome gains and losses and for hyperdiploidy (observed in 3 B-ALLs in this series). Moreover, Bionano identified also hyperdiploidy in a case with a normal karyotype. Interestingly, FISH analysis of the same case suggested the presence of a hypodiploid clone. This demonstrates the limitation of DNA based techniques to accurately determine ploidy when allele information is not available.
There were some discrepancies between the results of cytogenetics and Bionano:
Bionano could not identify the presence of multiple clones, a common limitation for DNA based techniques.
Bionano did not confirm some abnormalities identified by karyotype. Chromosome analysis of ALL samples is notoriously difficult due to low chromosome resolution explaining some of these differences. Some of the discrepant abnormalities were often marked as uncertain or were present only in a subclone (<10%).
Bionano allowed a correction of the karyotype in some cases and a refinement of the breakpoints of structural abnormalities.
Genome imaging revealed several additional intra- and inter-chromosomal translocations including (clonal) rearrangements of the TCR or IGH.
Overall our data demonstrate that genome imaging is a promising technique to identify structural and numerical abnormalities in ALL. Bionano provided an informative result in each case and results were concordant with other results in the majority of cases.
However this study also highlighted a few limitations. Further studies are underway to understand why the STIL-TAL1 was overlooked and to determine whether some of the additional variants identified by Bionano represent true rearrangements or technical artefacts.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal