Abstract
Until recently, the nucleic acid content of platelets was considered to be fully determined by their progenitor megakaryocyte. However, it is now well understood that additional mediators (eg, cancer cells) can intervene, thereby influencing the RNA repertoire of platelets. Platelets are highly dynamic cells that are able to communicate and influence their environment. For instance, platelets have been involved in various steps of cancer development and progression by supporting tumor growth, survival, and dissemination. Cancer cells can directly and/or indirectly influence platelet RNA content, resulting in tumor-mediated “education” of platelets. Alterations in the tumor-educated platelet RNA profile have been described as a novel source of potential biomarkers. Individual platelet RNA biomarkers as well as complex RNA signatures may be used for early detection of cancer and treatment monitoring. Here, we review the RNA transfer occurring between cancer cells and platelets. We explore the potential use of platelet RNA biomarkers as a liquid biopsy biosource and discuss methods to evaluate the transcriptomic content of platelets.
Platelets and cancer progression
Platelets were first described in 1865 as “colorless little spherules (…) 6-8 times smaller than the red cells.”1 A few years later, in 1882, Giulio Bizzozero comprehensively described these blood elements and named them as piastrine in Italian, meaning “small plates.” He conducted several biological experiments that have shown that platelets are anucleate cells and the first component of blood to adhere to damaged blood vessel walls in vitro and in vivo.1 Since then, many exciting studies have been performed indicating that the primary function of platelets is the regulation of hemostasis and thrombosis.2 Platelets originate from megakaryocytes present in the bone marrow and the lung through a systematic and regulated process. They remain in the bloodstream for an average period of 5 to 7 days.2,3 Although platelets are the smallest cells in blood circulation, they have an extraordinary capacity for morphological and internal changes, making them a key player in many pathophysiological processes. These processes include inflammation, atherogenesis, antimicrobial host defense, and tumor growth and metastasis.4
A large number of studies show a strong interaction between platelets and neoplastic cells. This association has been known since 1865, when Armand Trousseau first described that cancer cells can induce thrombosis formation (from here, the name Trousseau’s syndrome).5 Thrombosis is now considered a common clinical manifestation in cancer, with enhanced thrombus formation after tumor-induced platelet activation. Cancer cells can drive platelet activation through both direct and indirect mechanisms stimulating platelet aggregation (Figure 1), leading to a phenomenon called tumor cell–induced platelet aggregation (TCIPA).6 At least 2 pathways are involved in platelet activation during TCIPA. One is prompted by CLEC-2 and GPVI and induces a cascade of tyrosine phosphorylation downstream of the immune tyrosine activation motif.7-9 The second pathway involves TXA2, thrombin, and adenosine 5′-diphosphate, which interact with G protein–coupled receptors and initiate specific downstream signaling cascade.10-14 TCIPA is not only correlated with a high risk of thrombosis but also considered to promote angiogenesis and metastasis and therefore to negatively correlate with prognosis and survival.6,15,16
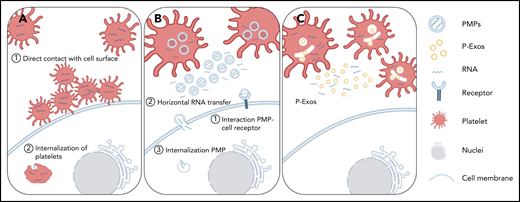
Schematic representation of direct and indirect mechanisms of platelet RNA transfer to nucleated cells. (A) Platelets can transfer part of their content to a recipient cell through direct contact with the membrane system of the target cell, or they can be internalized by the recipient cell. (B-C) Platelets can indirectly transfer their content by releasing PMPs or P-Exos. PMPs affect cellular responses through activation of cell surface receptors, horizontal transfer of RNA in and/or on PMPs through transient interaction with cell membranes, or internalization of PMPs and release of their content directly inside the recipient cell.
Schematic representation of direct and indirect mechanisms of platelet RNA transfer to nucleated cells. (A) Platelets can transfer part of their content to a recipient cell through direct contact with the membrane system of the target cell, or they can be internalized by the recipient cell. (B-C) Platelets can indirectly transfer their content by releasing PMPs or P-Exos. PMPs affect cellular responses through activation of cell surface receptors, horizontal transfer of RNA in and/or on PMPs through transient interaction with cell membranes, or internalization of PMPs and release of their content directly inside the recipient cell.
Interactions between tumor cells and platelets play an important role during all phases of cancer progression. Most of the interactions involve the recruitment of activated platelets that support cancer progression by facilitating tumor growth, angiogenesis, and metastasis. Platelets are considered to be part of the tumor microenvironment, which consist of not only cancer cells but also other cell types, such as immune cells and endothelial cells.17,18 Tumor cells can activate and recruit platelets by releasing tumor microparticles containing tissue factor and P-selectin glycoprotein ligand 1 (PSGL1).19 Another study demonstrated that platelets are actively recruited by colorectal tumor cells in a P-selectin–dependent manner. Once the platelets arrive in the tumor microenvironment, they adhere to the tumor cell membranes and induce the secretion of vascular endothelial growth factor (VEGF), which in turn promotes cell proliferation and angiogenesis.20 Activated platelets also produce transforming growth factor β (TGF-β), a potent immunosuppressive cytokine that generates a favorable tumor microenvironment. Tumor cells induce platelets to release TGF-β in order to escape from immune system recognition.21-24 For instance, TGF-β is implicated in the transformation of neutrophils into a protumorigenic phenotype, which impairs the immunosurveillance role of natural killer cells.21,25
Neutrophils in the tumor microenvironment were also described to form neutrophil extracellular traps (NETs).26 During the NETosis event, activated neutrophils externalize their DNA (both nuclear and mitochondrial) and granular content. It results in a 3-dimensional meshwork composed of decondensed chromatin and proteins, including myeloperoxidase, neutrophil elastase (NE) and LL37, that form an extracellular trap that is able to capture and kill bacterial and other pathogens.27,28 Decondensation and change of the charge of the chromatin occur during or prior to the NETosis event through an enzyme named peptidylarginine deiminase 4 (PAD4), which can lead to histone citrullination.29 Citrullinated histone H3 (H3Cit) has been identified to be a crucial marker of NET formation. High levels of H3Cit have been observed in the plasma of cancer patients and are associated with poor prognosis.30 Moreover, the presence of H3Cit in plasma of cancer patients is also correlated with high levels of other neutrophil activation markers, such as NE and myeloperoxidase. NETs have been identified as an important factor in tumor progression, metastasis, and cancer-associated thrombosis.31 Tumor-activated platelets have been suggested as a pro-NETosis component in the recruitment of neutrophils present in the tumor microenvironment.27,31,32 Likewise, the extracellular DNA present in the NETs seems to influence the activation of platelets and their prothrombotic function by enhancing TCIPA formation.28,33-35 In this manner, a self-sustaining process is initiated during which activated platelets can promote NET formation, and the NET content, conversely, can stimulate platelet activation.16,28 Moreover, cancer cells can exploit NETs in circulation. NETs interact with circulating tumor cells through β1-integrins present both in cancer cells and NETs.36 In this way, NETs facilitate the arrest and adhesion to distant organs of circulating tumor cells (CTCs).37 NETs also play a role in activation of the immune system through Toll-like receptor 9 expressed in immune cells as monocytes and dendritic cells. DNA in complex with LL-37 can also stimulate dendritic cells to produce proinflammatory interferon.38 Thus, NETs can stimulate the release of cytokines, leading to the activation of T cells and amplification of immune cell recruitment.39 Although it is well known that NETs play an important role in activation of the immune system, at the moment, their function in the immune response in the context of tumors is not fully understood. A study suggested a possible defensive role of NETs derived from tumor-associated neutrophils (TAMs) in tumor immunoediting.40 Another interesting study showed that NETs can influence the malignant transformation of B cells and enhance their proliferation in the context of autoimmune disease.41
Interaction between cancer cells and platelets also stimulates the production of platelet-derived angiogenic regulators.20 High levels of platelet-activated markers (such as P-selectin) and angiogenic markers (such as VEGF, angiopoietin, and platelet-derived growth factor) have been found in various types of cancer.42-44 Multiple signaling pathways are involved in the proangiogenic activity of platelets, but all of them are considered to implicate the action of cyclooxygenase.44 Cyclooxygenase inhibitors, such as aspirin, have been suggested to inhibit the angiogenic process in tumors by decreasing platelet-derived angiogenic proteins.45 Several studies suggest that chronic use of aspirin has an anticancer effect, thereby reducing tumorigenesis and distant metastasis formations (reviewed in Gresele et al46 ), although further research in this area is warranted. Purinergic receptors inhibitors, such as ticagrelor and clopidogrel, have shown to have antitumor and antimetastatic effects similar to aspirin. Cho et al have compared the antitumor effect of aspirin with ticagrelor, an inhibitor of P2Y12 receptor, in a murine model of ovarian cancer. Ticagrelor reduced tumor growth by 60% compared with aspirin.47 Similarly, clopidogrel, an irreversible P2Y12 inhibitor, can reduce tumor progression and help prevent thrombosis formation.48 These studies also reported an antimetastatic effect of these 2 compounds, which was correlated with inhibition of the purinergic receptors. Taken together, we can conclude that anticoagulant and antiplatelet drugs might be effective to prevent tumor progression and metastasis, but further clinical studies are needed before they can be used for cancer therapy.47-49
During the metastatic process, tumor cells detach from the primary tumor and enter the bloodstream. Before that, neoplastic cells can undergo epithelial-mesenchymal transition (EMT), which consists of disruption of cell-cell interactions, expression of mesenchymal markers, and increased mobility.50 Platelet-cancer cell interaction and platelet-derived TGF-β can synergistically activate the TGF-β/Smad and NF-κB pathways in cancer cells, promoting EMT in vivo.51 Moreover, activated platelets are a primary source of lysophosphatidic acid. This lipid, with growth factor signaling properties, upregulates the activity of matrix metalloproteinase 2 (MMP2), MMP7, and MMP9 in cancer cells. MMPs can mediate cancer progression by supporting cancer cell detachment from the primary site and their entry into the circulation.52 Once cancer cells have escaped from their microenvironment and enter the blood circulation, they are exposed to high shear stress and attack by the immune system. Without protection, <0.1% of cancer cells in the blood circulation will survive.53 In this phase, platelets can again play a key role in cancer cell protection. One process used by cancer cells to escape immunosurveillance is to mimic platelet physiology by expressing receptors and adhesion molecules present on the surface of platelets.54 Activated platelets are also able to transfer major histocompatibility complexes onto CTCs, thereby allowing cancer cells to be recognized as normal cells by the immune system.55 Moreover, during TCIPA, platelets express many adhesion molecules, including integrins and selectins, which enable platelets to attach to CTCs. This can create a physical shield that protects cancer cells from blood shear force and natural killer cells.31,56,57 The platelet-CTC complex can also promote tumor cell arrest and adhesion to the endothelial wall of blood vessels.51,58
Tumor cells need a specific environment with favorable conditions to grow and metastasize. Several factors help the formation of these optimal environment conditions, including platelet-derived signals. Platelets sequester different tumor-derived proteins, such as TGF-β1 and MMP1, which can support the formation of the prometastatic niche in the bone.59 In addition, Labelle et al demonstrated that platelets in contact with tumor cells release CXCL5 and CXCL7 chemokines, which can result in recruitment of granulocytes. These platelet-recruited granulocytes may lead to early metastatic niche formation.60
Platelet and tumor RNA cross talk
Despite their small size and lack of nuclei, platelets contain an ample repertoire of biomolecules, including functional ribosomes, signaling proteins, and different types of RNA, such as messenger RNA (mRNA), microRNA (miRNA), and circular RNA (circRNA). Previously, platelet content was considered to consist of “remnants” of progenitor megakaryocytes. Currently, we can appreciate that megakaryocytes specifically sort RNA transcripts into platelets.61 As demonstrated by the work of Cecchetti et al, megakaryocytes use regulated mechanisms to transfer specific MMP and TIMP mRNAs into newly generated platelets.61 Interestingly, the platelet transcriptome does not completely reflect its parental megakaryocyte profile. Analysis of the platelet proteome revealed that only 70% to 90% of the platelet transcriptome is represented.62,63 Platelets are able to actively respond to local and systematic conditions altering their transcriptome. In particular, alteration of the transcriptome profiles can occur under pathological conditions, including cancer.64,65 Due to their dynamic membrane, platelets can sequester RNA or modify their nucleic acid content in response to external stimuli. The ingestion of RNA by platelets can be considered as a random process, as no evidence of selective uptake of certain RNA transcripts has been identified. Conversely, platelets can also transfer functional RNA to recipient cells.66-68
Platelets have a unique membrane that allows the passage of small molecules and the transfer of their cytoplasmatic content to other cells. This unique structure permits several forms of communication through both direct (passage through the membrane system) and indirect (through vesicles) mechanisms.69 Risitano et al were the first to test the hypothesis that platelet-derived RNAs are implicated in cell-cell communication by acting as a source of transcripts. They developed an in vitro model using platelet-like particles, which have characteristics similar to human platelets. Functional cytosolic mRNAs were transferred into target cells through direct contact. This new communication model suggests that the platelet transcriptome is able to influence the gene expression of recipient cells.66 Subsequently, Kirschbaum et al observed the internalization of platelets by hepatocytes. Platelet-derived mRNAs were then released and translated into proteins by the recipient cells. This direct mechanism of RNA transfer promotes hepatocyte proliferation, and it may contribute to liver regeneration.70
Platelets can indirectly transfer their contents by releasing 2 types of membrane vesicles, microparticles (100 nm to 1 μm) and exosomes (40 to 100 nm).71 Platelet-derived microparticles (PMPs) represent the most abundant type of extracellular vesicles (EVs) in the body. They work as a biological vector for cell-cell communication.72,73 PMPs mediate intracellular signaling through 2 main mechanisms: (1) by acting as circulating signaling modules affecting cellular proprieties and responses through activation of receptors present on the surface of target cells, or 2) by transferring their internal content to recipient cells. This last transfer mechanism can occur by transient interaction, such as membrane association or internalization of PMPs into target cells.74 An increase in the number of circulating EVs has been observed in many types of cancer.75-78 The concentration of circulating PMPs has been shown to increase with cancer stage and therefore can be potentially used as a cancer biomarker.72 PMPs have been associated with many aspects of cancer progression, including angiogenesis, tumor growth, metastasis, escape from apoptosis and immune surveillance, extracellular matrix degradation, and chemoresistance.79,80 The first evidence that platelets can transfer functional RNA through PMPs to endothelial cells was demonstrated by Gidlӧf et al and Laffont et al. Gidlӧf et al observed that activated platelets can transfer 4 types of miRNA (miR-22, miR-185, miR-320b, and miR-423-5p) into endothelial cells and thereby regulate the expression of their target genes.68 Similarly, Laffont et al showed degradation of the mRNA target due to the transfer of platelet-derived Ago2-miR-223 complexes.67 Liang et al reported high levels of miR-223 both in platelets and in PMPs of non-small cell lung cancer (NSCLC) patients as compared with healthy controls. They further observed that miR-223 is efficiently delivered into cancer cells through PMPs, leading to the downregulation of one of its targets, EPB41L3, and consequently promoting cell invasion.81 The miRNA Let-7a in PMPs can instead promote angiogenesis by leading to the reduction of the antiangiogenic THB-1 in endothelial cells.82 Furthermore, miR-126-3p can be delivered into macrophages, altering their gene expression and reprogramming their immunosurveillance function.83 Michael et al showed that PMPs can also have an antitumorigenic function. In an in vivo study, miR-24 transferred into tumor cells led to tumor cell apoptosis.84 PMPs can also transfer other types of RNA. Yao et al observed a high level of TPM3 mRNA in platelets from breast cancer patients, which significantly correlated with high metastatic risk. They reported that TPM3 mRNA is delivered to cancer cells via PMPs, thereby promoting cancer cell migration.85 Similarly, Yang et al observed that TIMP1 mRNA can be carried into colorectal cancer cells, where it can promote tumor growth in vitro and in vivo.86 Recently, Plantureux et al investigated platelet-educated cancer cells.87 They observed that interaction of platelets with tumor cells through cadherin-6 can lead to the production of platelet- and tumor cell–interacting microparticles (iMPs).87 At the primary tumor site, iMPs can recruit and activate macrophages that promote cell-proliferation arrest through interferon-γ and interleukin-4. In contrast, platelets and iMPs in the bloodstream can instead promote metastasis by inducing EMT and interaction between cancer cells and endothelial cells, thereby changing the expression level of mRNAs involved in inflammation and metastasis in cancer cells.87
The second type of vesicles released by platelets are exosomes (P-Exos). They are generated from multivesicular bodies and α-granules.71 P-Exos are highly enriched in CD41 in addition to other classical exosome markers.88 P-Exos are supplied with specific miRNAs, such as miR-21, miR-223, miR-339, and miR-328.89-91 Exosomes are involved in several steps of cancer progression, including angiogenesis, cell migration and invasion, immunosuppression, and formation of the metastatic niche.92 Although there are numerous studies regarding exosomes derived from cancer cells, we have limited information on the role of P-Exos during cancer progression. Further studies are needed to elucidate the impact of these processes.
Cancer cells “educate” platelet transcriptomes
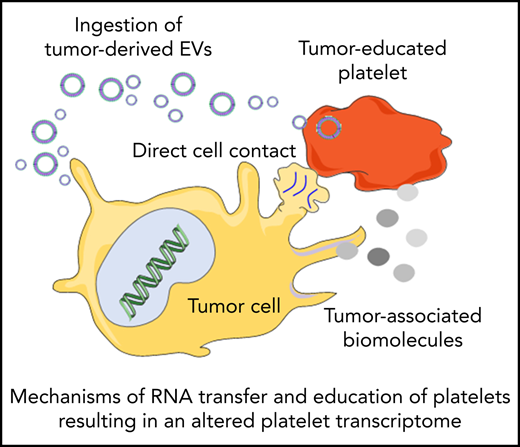
Platelets may constantly receive stimuli and sequester biomolecules coming from other cells, leading to alterations of the platelet transcriptome.93 This can occur through direct contact with the cells by membrane proteins or through extracellular molecules released by the cells. Interactions between platelets and cancer cells takes place through several adhesion molecules, including P-selectin, integrins, and glycoproteins present on the platelet membrane, leading to platelet responses and activation.94 On the other hand tumor cells can also release extracellular metabolites, such as proteinases (in particular thrombin), tissue factors, adenosine 5′-diphosphate, MMPs, and TXA2, that can activate platelet receptors. These molecules can be released directly in the circulation or packaged in tumor-derived EVs. EVs can also contain tumor-derived RNAs, which can be transferred to platelets.95 Tumor cells may exploit these mechanisms to educate platelets, which could result in alteration of the platelet RNA content in response to cancer cell signals. This can occur in at least 3 ways: (1) induction of protein translation and subsequent RNA decay, (2) stimulation of specific RNA splicing events, and (3) sequestration and release of RNA in circulation. De novo protein synthesis in platelets was discovered >50 years ago.96 External stimuli, such as lipopolysaccharide, P-selectin, and thrombin, can activate protein synthesis in platelets.97-101 mRNA translation can be also modulated by alteration of the miRNA repertoire in platelets.102 Thus, tumor cells may directly (through transfer of tumor-derived RNA) and indirectly (through releasing signals that modulate platelet mRNA processing) stimulate protein synthesis in platelets, resulting in protumorigenic platelets.103-105 Platelets contain functional spliceosomes that are able to splice pre-mRNA upon signals in activated platelets.106 Alterations of spliceosome function in cancer may be one of the causes of differential spliced events observed in the tumor-educated platelet (TEP) RNA signature profile.64,65,107 Furthermore, platelets are able to sequester spliced and unspliced mRNA from cancer cells.10,95,108 Since platelets do not contain a nucleus, they cannot produce new transcripts. Platelets can acquire new transcripts via ingestion and are also reported to preserve the megakaryocyte-derived transcripts. Hence, platelets may be equipped to protect accumulating tumor-derived RNA transcripts. However, as indicated in Table 1, platelet biomarker transcripts can be transferred from tumor cells to platelets, although in many of the listed studies, the putative origin of the biomarker transcripts is the megakaryocyte (endogenous). These data are indicative of a highly dynamic RNA repertoire in platelets, potentially providing a unique biomarker resource for blood-based liquid biopsies. Currently, clinical oncology practice relies on tumor tissue biopsies for cancer diagnostics. Tissue biopsy is an invasive practice that can be accompanied by surgical complication and pain; for this reason, it cannot be repeated often. In many occasions, it is also not possible to take a tissue biopsy specimen containing high-quality tumor material. Liquid biopsies offer a minimally invasive and safe alternative or complimentary approach to tissue biopsies, making possible multiple sampling. Liquid biopsies not only enable the detection of tumors but can also be relevant for prognosis, personalized treatment, monitoring of the therapy, and prediction of recurrences. In the last few years, an increasing number of studies have shown the potential of TEPs as a liquid biopsy biosource.104
Overview of TEP mRNA cancer biomarker studies
| mRNA TEP biomarkers . | Cancer type . | Findings . | Putative predominant biomarker origin . | Detection method . | References . | Predictive value . |
|---|---|---|---|---|---|---|
| Panel of mRNAs | NSCLC | Detection of metastatic lung cancer | Endogenous | Microarray | 108 | 200 transcripts DE between NSCLC and controls with P < 1 × 10−4 |
| EGFRvIII and PCA3 | Glioma and PCa | Tumor-derived (mutant) RNAs are transferred into platelets | Transferred | RT-PCR | 95 | EGFRvIII present in 80% of the platelets derived from patients who are EGFRvIII positive |
| EML4-ALK | NSCLC | Biomarker for monitoring of chemotherapy response (crizotinib) | Transferred | qRT-PCR | 110 | RT-PCR demonstrated 65% sensitivity and 100% specificity for the detection of EML4-ALK rearrangements in platelets |
| Panel of mRNAs | NSCLC, CRC, GBM, BrCa, PAAD, HBC | Cancer detection and identification location primary tumor. Identification of MET or HER2-positive, and mutant KRAS, EGFR, or PIK3CA tumors | Endogenous | RNA-seq | 65 | 96% accuracy to distinguish patients with localized and metastasized tumors from healthy individuals; 71% accuracy to identify primary tumor location across 6 different tumor types |
| KLK2, KLK3, FOLH1, and NPY | PCa | Biomarkers for treatment response (abiraterone) | Transferred | ddPCR | 112 | Combining KLK3, FOLH1, and NPY separates long-term responders from short-term responders with 87% sensitivity and 82% specificity |
| Panel of mRNAs | NSCLC | Detection of early- and late-stage NSCLC | Endogenous | RNA-seq | 64 | Accuracy 88%, AUC 0.94, 95% CI 0.92-0.96, P < .001 for detection of late-stage NSCLC; accuracy 81%, AUC 0.89, 95% CI 0.83-0.95, P < .001 for detection of early-stage NSCLC |
| Panel of mRNAs | NSCLC | Early detection of NSCLC | Endogenous | RNA-seq | 125 | Sensitivity 0.925, specificity 0.827, accuracy 0.889, and Matthews correlation coefficients 0.760 for detection of NSCLC |
| EGFRvIII | NSCLC | Biomarker for identification of appropriate treatment selection | Transferred | RT-PCR | 109 | EGFRvIII was detected in 1.5% of all patients analyzed |
| Panel of mRNAs | MM | Transcripts involved in platelet activation, signaling, and aggregation were upregulated in SMM/MM patient platelets compared with HD platelets | Endogenous | RNA-seq | 23 | Panel of 336 transcripts increased and 441 transcripts decreased |
| ALK rearrangement | NSCLC | Diagnosis of ALK-positive NSCLC and predictive value for ALK inhibitors treatment | Transferred | RT-PCR | 111 | Accuracy 80.3% for detection of ALK rearrangements using platelets |
| ITGA2B | NSCLC | Identification of stage I NSCLC and differentiate malignant from benign lung nodules | Endogenous | RNA-seq, qRT-PCR, and ddPCR | 91 | AUC 0.888, sensitivity 91.2%, and specificity 56.5% for detection of NSCLC by qRT-PCR |
| MAX, MTURN, HLA-B | Lung cancer | Detection of early-stage lung cancer patients and predictive response of chemotherapy treatment | Endogenous | Microarray, qRT-PCR | 117 | AUC 0.734, sensitivity of 60.6%, specificity of 81.7% for detection of lung cancer patients compared with healthy controls |
| AUC 0.787, sensitivity of 72.7%, specificity of 85.4%, for detection in early-stage lung cancer compared healthy donors | ||||||
| TIMP1 | CRC | TIMP1 is expressed at high levels in CRC patients; TIMP1 mRNA can be transferred to CRC cells and promote tumor growth in vivo and in vitro | Transferred | RNA-seq and qRT-PCR | 86 | AUC 0.9583 (95%CI, 0.9363–0.9803) |
| TPM3 | BrCa | TPM3 is present at high level in patients with BrCa; TPM3 mRNA is transferred through microvescicles and enhances cancer cell migration | Transferred | RNA-seq and qRT-PCR | 85 | AUC 0.9705 (95% CI, 0.9494-0.9823) for detection of BrCa |
| AUC 0.8404 (95% CI, 0.7566-0.9242) for detection of metastatic BrCa | ||||||
| Panel of mRNAs | Sarcoma | Diagnosis of sarcoma patients | Endogenous | RNA-seq | 126 | Accuracy of 87%, AUC 0.93 (95% CI, 0.86-1) |
| mRNA TEP biomarkers . | Cancer type . | Findings . | Putative predominant biomarker origin . | Detection method . | References . | Predictive value . |
|---|---|---|---|---|---|---|
| Panel of mRNAs | NSCLC | Detection of metastatic lung cancer | Endogenous | Microarray | 108 | 200 transcripts DE between NSCLC and controls with P < 1 × 10−4 |
| EGFRvIII and PCA3 | Glioma and PCa | Tumor-derived (mutant) RNAs are transferred into platelets | Transferred | RT-PCR | 95 | EGFRvIII present in 80% of the platelets derived from patients who are EGFRvIII positive |
| EML4-ALK | NSCLC | Biomarker for monitoring of chemotherapy response (crizotinib) | Transferred | qRT-PCR | 110 | RT-PCR demonstrated 65% sensitivity and 100% specificity for the detection of EML4-ALK rearrangements in platelets |
| Panel of mRNAs | NSCLC, CRC, GBM, BrCa, PAAD, HBC | Cancer detection and identification location primary tumor. Identification of MET or HER2-positive, and mutant KRAS, EGFR, or PIK3CA tumors | Endogenous | RNA-seq | 65 | 96% accuracy to distinguish patients with localized and metastasized tumors from healthy individuals; 71% accuracy to identify primary tumor location across 6 different tumor types |
| KLK2, KLK3, FOLH1, and NPY | PCa | Biomarkers for treatment response (abiraterone) | Transferred | ddPCR | 112 | Combining KLK3, FOLH1, and NPY separates long-term responders from short-term responders with 87% sensitivity and 82% specificity |
| Panel of mRNAs | NSCLC | Detection of early- and late-stage NSCLC | Endogenous | RNA-seq | 64 | Accuracy 88%, AUC 0.94, 95% CI 0.92-0.96, P < .001 for detection of late-stage NSCLC; accuracy 81%, AUC 0.89, 95% CI 0.83-0.95, P < .001 for detection of early-stage NSCLC |
| Panel of mRNAs | NSCLC | Early detection of NSCLC | Endogenous | RNA-seq | 125 | Sensitivity 0.925, specificity 0.827, accuracy 0.889, and Matthews correlation coefficients 0.760 for detection of NSCLC |
| EGFRvIII | NSCLC | Biomarker for identification of appropriate treatment selection | Transferred | RT-PCR | 109 | EGFRvIII was detected in 1.5% of all patients analyzed |
| Panel of mRNAs | MM | Transcripts involved in platelet activation, signaling, and aggregation were upregulated in SMM/MM patient platelets compared with HD platelets | Endogenous | RNA-seq | 23 | Panel of 336 transcripts increased and 441 transcripts decreased |
| ALK rearrangement | NSCLC | Diagnosis of ALK-positive NSCLC and predictive value for ALK inhibitors treatment | Transferred | RT-PCR | 111 | Accuracy 80.3% for detection of ALK rearrangements using platelets |
| ITGA2B | NSCLC | Identification of stage I NSCLC and differentiate malignant from benign lung nodules | Endogenous | RNA-seq, qRT-PCR, and ddPCR | 91 | AUC 0.888, sensitivity 91.2%, and specificity 56.5% for detection of NSCLC by qRT-PCR |
| MAX, MTURN, HLA-B | Lung cancer | Detection of early-stage lung cancer patients and predictive response of chemotherapy treatment | Endogenous | Microarray, qRT-PCR | 117 | AUC 0.734, sensitivity of 60.6%, specificity of 81.7% for detection of lung cancer patients compared with healthy controls |
| AUC 0.787, sensitivity of 72.7%, specificity of 85.4%, for detection in early-stage lung cancer compared healthy donors | ||||||
| TIMP1 | CRC | TIMP1 is expressed at high levels in CRC patients; TIMP1 mRNA can be transferred to CRC cells and promote tumor growth in vivo and in vitro | Transferred | RNA-seq and qRT-PCR | 86 | AUC 0.9583 (95%CI, 0.9363–0.9803) |
| TPM3 | BrCa | TPM3 is present at high level in patients with BrCa; TPM3 mRNA is transferred through microvescicles and enhances cancer cell migration | Transferred | RNA-seq and qRT-PCR | 85 | AUC 0.9705 (95% CI, 0.9494-0.9823) for detection of BrCa |
| AUC 0.8404 (95% CI, 0.7566-0.9242) for detection of metastatic BrCa | ||||||
| Panel of mRNAs | Sarcoma | Diagnosis of sarcoma patients | Endogenous | RNA-seq | 126 | Accuracy of 87%, AUC 0.93 (95% CI, 0.86-1) |
AUC, area under the curve; BrCa, breast cancer; CI, confidence interval; CRC, colorectal cancer; DE, differentially expressed; GBM, glioblastoma; HBC, hepatobiliary cancer; HD, healthy donor; MM, multiple myeloma; PAAD, pancreatic adenocarcinoma; PCa, prostate cancer; RNA-seq, RNA sequencing; SMM, smoldering multiple myeloma.
TEP RNA as biosource for blood-based liquid biopsy
A first insight in the diagnostic TEP transcriptome potential was given by Calverley et al in 2010. They profiled platelet mRNA from metastatic lung cancer patients and healthy donors using microarrays and identified altered expression levels of 200 RNAs and 608 altered splicing events. One of these differentially spliced mRNAs is the NAD-dependent deacetylase sirtuin-2 (SIRT2), which plays a role in epigenetic silencing. It was suggested that platelets could induce tumor growth and progression by releasing epigenetic silencers.108 Subsequently, Nilsson et al showed that platelets from glioma patients have a distinct RNA signature as compared with healthy controls. Moreover, they demonstrated that tumor cells can transfer (mutant) RNA into platelets via tumor-derived EVs. Tumor-derived epidermal growth factor receptor (EGFR) vIII and PCA3 transcripts were identified in platelets isolated from glioma and prostate cancer patients, respectively.95 In another study. EGFRvIII was also detected in platelets derived from NSCLC patients.109 Platelets derived from patients with cancer can carry fusion transcripts derived from the tumor, as was demonstrated for EML4-ALK in platelets isolated from lung cancer patients. The monitoring of EML4-ALK–rearranged fusion transcripts may be of particular importance for the prediction of treatment responses to ALK inhibitors in patients with lung cancer.110 The ALK-rearrangement liquid biopsy test could successfully predict the treatment outcome of ALK inhibitors in patients.111 That platelets can contain predictive therapy response biomarkers was confirmed in another study on castration-resistant prostate cancer. By combining tumor-associated RNA biomarkers (KLK3, KLK2, FOLH1, and NPY), it was feasible to predict the outcome of abiraterone therapy in those patients.112 It is unlikely that ingestion of RNA transcripts present in a single EV can be responsible for significant changes of the platelet transcriptome, although RNA transferred by multiple EVs may accumulate to significant levels in platelets, as demonstrated for EGFRvIII95 and EML4-ALK.110 It also seems unlikely that tumor-derived RNA delivered via EVs contributes significantly to the altered platelet transcriptome as measured by methods such as shallow sequencing, since deep sequencing or sensitive reverse transcription polymerase chain reaction (RT-PCR) methods were required to detect tumor-derived transcripts such as EGFRvIII and EML4-ALK. More research is warranted to study the effect of EVs on the induction of RNA splicing in platelets as well as RNA expression and sorting in megakaryocytes. Moreover, based on current knowledge, we cannot conclusively identify the exact origin of the platelet biomarker transcript. Additional track-and-trace studies are warranted to demonstrate the transfer of transcripts from tumor cells to platelets in order to convincingly demonstrate the origin of the biomarker transcript. One study used the transcriptome profile of platelets to develop a predictive algorithm to efficiently separate patients from healthy individuals. In this work, platelet transcriptomes were isolated from blood collected in EDTA tubes, which were developed to bring platelets in a stable coma-like state. During the first 72 hours after blood collection in EDTA tubes, only limited changes in the platelet RNA profile are measured in the presence of EDTA.64,107 The relative stability of platelet RNA in the absence of stimuli was also demonstrated by Rondina et al, who found a remarkable stability of platelet RNA in individuals over time.113 In this way, they ensured that the differences in the transcriptome profile are not due to preanalytical variables. Nevertheless, stability needs to be demonstrated for any protocol used. Support vector machine classification based on mRNA platelet profiles allowed them to correctly classify cancer patients with an accuracy of 96% and identify the location of the primary tumor among 6 cancer types with an accuracy of 71%. Specific spliced RNA signatures were identified to be associated with tumor tissue molecular subtype and enabled their identification with an accuracy of 85% to 95%. Surrogate TEP RNA profiles allow them to distinguish patients with KRAS and EGRF mutations in their tumor tissue, overexpression of MET, and patients with breast cancer having HER2-amplified, PIK3CA mutant, or triple-negative tumor tissues. The platelet surrogate signatures were developed as an all-in-one readout system to identify multiple tumor tissue mutations from a single thromboSeq platelet sequencing file using mutation-specific classifiers, thereby avoiding the need to multiplex different targeted assays to detect a panel of mutants. The altered transcripts of the surrogate signatures are thus an indirect method of measuring mutations. It is likely that most of the spliced transcripts of the surrogate signatures are not tumor derived but rather are transcripts derived from megakaryocytes that are altered in response to tumor-associated signals. Such transcript alteration may already occur in the megakaryocyte before sorting of the transcripts into platelets, or this may happen inside the platelets during circulation or once in contact with tumor cells. To evaluate the efficiency of mutation detection of surrogate TEP RNA signatures, a comparison was made to the use of targeted amplicon deep sequencing (AmpliSeq) for KRAS and EGFR mutations. It was observed that surrogate TEP RNA signatures by shallow sequencing can provide a relatively high diagnostic accuracy (81%, P < .05), although the targeted deep sequencing AmpliSeq approach also resulted in the detection of mutant transcripts, albeit at significantly lower accuracy (36% to 39%, P < .05). The AmpliSeq method also was significantly less accurate than the corresponding measurements for plasma (circulating free DNA), suggesting that plasma has a better wild-type (WT)/mutant ratio for the detection of certain mutant tumor-derived markers as compared with the platelet fraction.114 A recent study115 reports that platelets contain more WT BRAF transcripts than EVs isolated from plasma (680 000 WT BRAF transcripts in platelets vs 2,670 WT transcripts in the EV fraction). As a consequence, based on a reported mutant BRAF assay sensitivity of 0.01%, no less than 68 mutants can be detected in the platelet fraction. While on average ∼23 BRAF mutants could be detected in the plasma EV fraction (2670 WT × 0.856% mutant allele fraction), no BRAF mutants were detected in platelets, which is likely due to the large number of WT BRAF transcripts present in platelets in combination with the assay sensitivity. However, when comparing the detection of the “WT-independent” fusion marker EML4-ALK in plasma (ie, total plasma RNA fraction, including EV-derived RNA) to platelets, a strong advantage was shown for platelets (22/34 detected) vs plasma (3/14 detected).110 Hence, a potential important aspect when taking biosources and assay sensitivity into account is the WT/mutant ratio of the marker to be identified.
Several groups have investigated the use of platelet RNA profiles for the detection of cancer, which is summarized in Table 1 and reviewed elsewhere.116 Currently, studies using TEPs are mostly focused on the biomarker discovery phase. It is critical to design large prospective studies that are statistically powered to validate individual TEP markers or their RNA signatures for potential future clinical implementation.116
Emerging technologies for TEP biomarkers
The assessment of platelet transcriptome alterations during pathological conditions and exchange of RNAs molecules has sparked the use of a wide range of molecular technologies. Conventional high-throughput technologies have partially revealed the complexity of the transcriptome profiles of platelets. However, the detection of tumor-derived RNAs requires very sensitive techniques, such as ultra-deep massive parallel sequencing or targeted methods such as droplet digital PCR (ddPCR), RT-PCR, or amplification refractory mutation system RT-PCR. As pointed out in a recent publication,115 platelets can contain very high numbers of certain WT transcripts, which can complicate (and some cases make impossible) the detection of tumor-derived biomarkers. For this matter, an optimal biomarker-background ratio is necessary to ensure the identification and the development of tumor-biomarker detection assays. This also indicates that direct RNA transfer from cancer cells to platelets does not strongly contribute to the altered RNA profile upon education of the platelets as measured by shallow sequencing. It remains to be investigated if such transferred tumor-derived RNA can be translated in the platelets, thereby potentially contributing to the educational process. Enhancing sensitivity and accuracy of detection techniques and specific selection of TEPs will help to implement the use of TEPs as liquid biopsies for cancer detection and personalized therapy selection. Third-generation or long-read sequencing may be another potential method to improve the assessment of variations occurring in the transcriptome profile of platelets. This technology allows the sequencing of full-length transcripts that, besides simplifying the detection of transcript isoforms, supports the identification of novel splicing forms and transcripts that were not detectable by short-read sequencing. This new technology may lead to the recognition of new cancer biomarkers in TEPs. Additionally, Oxford Nanopore technology developed a protocol that permits the direct sequencing of RNA molecules without bias due to retrotranscription and RNA amplification. With this method, the RNA is sequenced in its native state, enabling the detection of epitranscriptomic base modifications.117-119 Alterations in the epitranscriptomic profile have been previously correlated with cancer,120 and such RNA modifications in the TEP transcriptome may represent a new type of diagnostic biomarker. Another potential biomarker identified in platelets is circRNA. They are generated from pre-mRNAs through a mechanism of back splicing. Platelets are 17- to 188-fold enriched in circRNAs compared with several types of nucleated cells.121 circRNAs can function as a sponge for miRNA and RNA-binding proteins. In this way, they modulate the transcriptome profile of the cell. There is evidence that open reading frames in the circRNA can be translated, thereby producing functional peptides.122 Alhasan et al hypothesized that circRNAs function as RNA storage backup to protect mRNA from decay. Moreover, circRNA can be selectively released by platelets into vesicles, and they may be involved in signaling pathways.123 Several studies showed that their expression is altered in different pathological conditions, including cancer.124 In conclusion, platelets are a source of RNA biomarkers due to their ability to exchange information and modulated their content in response to the environment. Further studies of RNA transfer mechanisms between platelets and cancer cells can help us to better understand and monitor cancer progression.
Acknowledgment
This work was funded by the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie grant agreement 765492.
Authorship
Contribution: S.D., R.J.N., and T.W. wrote the paper.
Conflict-of-interest disclosure: R.J.N. and T.W. are shareholders of Grail. S.D. declares no competing financial interests.
Correspondence: Thomas Wurdinger, VU University Medical Center, De Boelelaan 1118, 1081 HV Amsterdam, The Netherlands; e-mail: t.wurdinger@amsterdamumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal