Key Points
A diplotype spanning the coding region of the IFNL4 gene influences molecular response to IFN-α therapy in polycythemia vera.
Determining IFNL4 diplotype status rather than typing tagSNPs may allow for optimizing patient management during IFN-α treatment of polycythemia vera.
Abstract
Interferon-α (IFN-α)–based treatments can induce hematologic and molecular responses (HRs and MRs, respectively) in polycythemia vera (PV); however, patients do not respond equally. Germline genetic factors have been implicated in differential drug responses. We addressed the effect of common germline polymorphisms on HR and MR after treatment of PV in the PROUD-PV and CONTINUATION-PV studies in a total of 122 patients who received ropeginterferon alfa-2b. Genome-wide association studies using longitudinal data on HR and MR over a 36-month follow-up did not reveal any associations at the level of genome-wide statistical significance. Furthermore, we performed targeted association analyses at the interferon lambda 4 (IFNL4) locus, well known for its role in hepatitis C viral clearance and recently reported to influence HR during treatment of myeloproliferative neoplasms. We did not observe any association of IFNL4 polymorphisms with HR in our study cohort; however, we demonstrated a statistically significant effect of the functionally causative IFNL4 diplotype (haplotype pair, including the protein-coding variants rs368234815/rs117648444) on MR (P = 3.91 × 10−4; odds ratio, 10.80; 95% confidence interval, 2.39-69.97) as reflected in differential JAK2V617F mutational burden changes according to IFNL4 diplotype status. Stratification of patients with PV based on IFNL4 functionality may allow for optimizing patient management during IFN-α–based therapy.
Introduction
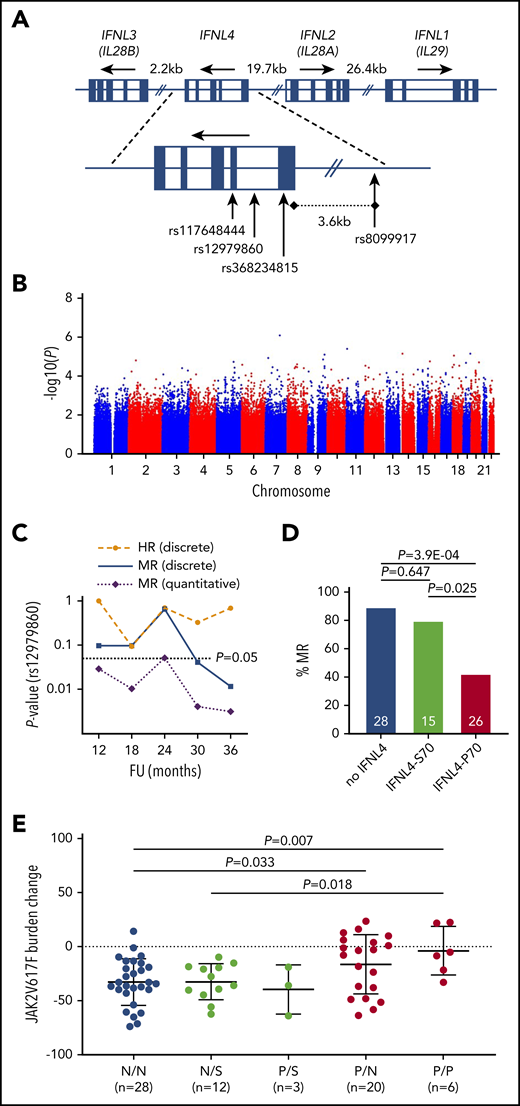
Interferon-α (IFN-α)–based therapies can induce sustained hematologic responses (HRs) and durable molecular responses (MRs) in polycythemia vera (PV) and other myeloproliferative neoplasms (MPNs).1 During treatment of PV, an HR reflects normalization of blood values, whereas a MR marks a substantial reduction of the malignant clone in the peripheral blood, when quantified by the allelic burden of the JAK2V617F mutation that drives the disease in 95% of patients with PV.1-4 IFN-α has been consistently reported to have a disease-modifying capacity whereby it selectively targets malignant cells, which induces durable MRs in some patients, whereas other patients respond insufficiently.5-7 Previous studies have suggested that certain features of disease-driving somatic mutations and genomic aberrations do not predict the response to treatment with IFN-α.6-10 The role of germline polymorphisms on a genome-wide scale has not been studied comprehensively in MPNs treated with IFN-α. In patients with hepatitis C, however, germline variation at the interferon lambda 4 (IFNL4) locus11,12 (Figure 1A) has been reported to strongly affect both spontaneous and IFN-α–induced viral clearance.13-15
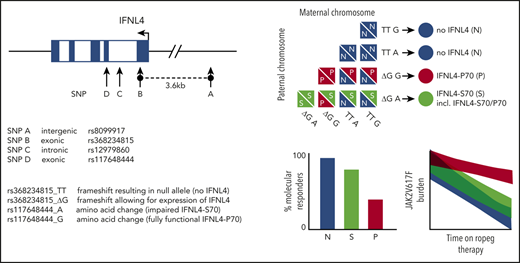
Germline genetic variation within the protein-coding region of IFNL4 affects MR during IFN-α treatment of PV. (A) Genomic organization of the IFNL locus on human chromosome 19q13.2. (B) An association plot derived from GWAS performed for MR after a 12-month follow-up of ropeg treatment (n = 102 patients). The plot is representative of all GWAS performed on both HR and MR data after a 12-, 18-, 24-, 30-, and 36-month follow-up. (C) Longitudinal P values for the association of the IFNL4 tagSNP rs12979860 with response to ropeg over a 36-month follow-up (FU). (D) Fraction (%) of MRs in the 3 main functional categories: patients who produced no functional IFNL4 (no IFNL4), those who produced impaired IFNL4-S70 (IFNL4-S70 alone+IFNL4-S70/P70), and those who produced fully functional IFNL4-P70 alone. Numbers within the bars represent total patients in the categories. (E) JAK2V617F mutant allele burden changes (absolute change from baseline value) at a 36-month follow-up in patients stratified according to IFNL4 diploid functional status. N, no IFNL4; S, IFNL4-S70; P, IFNL4-P70.
Germline genetic variation within the protein-coding region of IFNL4 affects MR during IFN-α treatment of PV. (A) Genomic organization of the IFNL locus on human chromosome 19q13.2. (B) An association plot derived from GWAS performed for MR after a 12-month follow-up of ropeg treatment (n = 102 patients). The plot is representative of all GWAS performed on both HR and MR data after a 12-, 18-, 24-, 30-, and 36-month follow-up. (C) Longitudinal P values for the association of the IFNL4 tagSNP rs12979860 with response to ropeg over a 36-month follow-up (FU). (D) Fraction (%) of MRs in the 3 main functional categories: patients who produced no functional IFNL4 (no IFNL4), those who produced impaired IFNL4-S70 (IFNL4-S70 alone+IFNL4-S70/P70), and those who produced fully functional IFNL4-P70 alone. Numbers within the bars represent total patients in the categories. (E) JAK2V617F mutant allele burden changes (absolute change from baseline value) at a 36-month follow-up in patients stratified according to IFNL4 diploid functional status. N, no IFNL4; S, IFNL4-S70; P, IFNL4-P70.
In this study, we addressed the effect of germline genetic factors on the outcome of treatment with ropeginterferon alfa-2b (ropeg), a monopegylated IFN-α, in a cohort of patients with PV (N = 122). We performed genome-wide association studies (GWAS) as an unbiased approach and additionally evaluated the potential influence of IFNL4 polymorphisms on HR and MR during treatment of PV.
Study design
Data on HR and MR were collected in patients with PV in the ropeg arm of the PROUD-PV (www.clinicaltrials.gov, NCT01949805) and CONTINUATION-PV (NCT02218047) clinical trials16 (N = 122). For replication, we included data on MR from the PEGINVERA (NCT01193699) study9 (n = 27). We restricted the HR evaluation to complete HRs only, whereas the MR analysis comprised both complete MRs and partial MRs, defined according to European LeukemiaNet criteria.17 Genome-wide genotypes were derived from Affymetrix SNP 6.0 microarrays. Additional IFNL4 variants were sequenced and subsequently phased with SHAPEIT.18 Association analyses were performed with PLINK19 and R.20 Details on study design and patient characteristics have been described elsewhere16 ; additional methods are detailed in the supplemental Information (available on the Blood Web site).
Results and discussion
To test for potential associations between germline genetic variation and response to IFN-α, we performed GWAS for HRs and MRs at a 12-, 18-, 24, 30-, and 36-month follow-up after ropeg treatment (supplemental Table 1). After standard quality controls, as implemented in GWAS (supplemental Figures 1 and 2), genome-wide tagging single-nucleotide polymorphisms (tagSNPs) were tested for allelic association in a case-control setup. For MR, we also applied a test to determine quantitative trait association, using changes in JAK2V617F mutational burden as continuous variables. None of those analyses revealed any association that reached genome-wide statistical significance after Bonferroni correction for multiple testing (ie, P < 5.00 × 10−8; Figure 1B). Although the statistical power of GWAS is limited for small sample numbers, a cohort of similar size has revealed variants that have a strong impact on IFN-α treatment of hepatitis C.13 Our results suggest that no germline factors influence IFN-α treatment outcome in PV with an effect size equivalent to that of IFN-α in hepatitis C. These results indicate that all patients with PV may be eligible for ropeg therapy, independent of their genetic makeup.
Although GWAS has the potential to agnostically identify associations, conservative correction for multiple testing limits the power of detecting true positive associations. This can be overcome by hypothesis-based targeted testing. Our targeted analysis focused on variants implicated in response to hepatitis C virus.13-15 Two such variants, specifically the noncoding SNPs rs8099917 and rs12979860, have been reported to affect HR during IFN-α treatment of a Swedish mixed PV and essential thrombocythemia cohort.21 To replicate those findings regarding HR and to investigate a potential effect on MR, we performed a series of association analyses for longitudinal time points up to the 36-month follow-up in our cohort. For hepatitis C virus clearance, Terczyńska-Dyla et al22 demonstrated that a diplotype (ie, a specific combination of 2 haplotypes) that includes 2 exonic IFNL4 variants covers most of the causality. Rs368234815_TT disrupts the open reading frame (IFNL4 loss of function; no IFNL4), whereas the rs117648444_G variant generates the impaired IFNL4-S70 protein in contrast to the fully functional IFNL4-P70 (Figure 1A). Interestingly, it is the functional IFNL4-P70 that exhibits a negative impact on viral clearance.22 For both variants, we included the individual genotypes, as well as the phased diplotypes into our analyses.
When testing our cohort for genotypic association of rs12979860 with HR and MR in a case-control setup, we could not replicate the previously reported findings on HR; however, for MR we observed a statistically significant association at a 36 months follow-up (P = .0065; odds ratio [OR], 6.10; 95% confidence interval [CI], 1.49-36.64; Figure 1C; Table 1). Notably, when testing for changes of JAK2V617F burdens as a quantitative trait, we found the association to be present at formal statistical significance at all stages of follow-up (Figure 1C), suggesting superior statistical power in testing quantitative burden as compared with discrete response categories defined on arbitrary thresholds.
Association of IFNL4 polymorphisms with HRs and MRs at 36-month follow-up of ropeg treatment
| Variant/diplotype . | Genotype/diplotype status . | n . | NR, n . | R, n . | % R . | OR (95% CI) . | P . |
|---|---|---|---|---|---|---|---|
| Influence of IFNL4 individual variants on HR in ropeg treatment of PV | |||||||
| rs8099917 | Non-TT | 29 | 8 | 21 | 72.4 | ||
| TT | 49 | 7 | 42 | 85.7 | 2.26 (0.62-8.45) | .2336 | |
| rs12979860 | Non-CC | 48 | 10 | 38 | 79.2 | ||
| CC | 31 | 5 | 26 | 83.9 | 1.36 (0.37-5.70) | .7710 | |
| rs368234815 | Non-TT | 48 | 10 | 38 | 79.2 | ||
| TT | 31 | 5 | 26 | 83.9 | 1.36 (0.37-5.70) | .7710 | |
| rs117648444 | GG | 58 | 13 | 45 | 77.6 | ||
| GA | 21 | 2 | 19 | 90.5 | 2.71 (0.53-27.11) | .3303 | |
| Influence of IFNL4 individual variants on MR in ropeg treatment of PV | |||||||
| rs8099917 | Non-T/T | 28 | 14 | 14 | 50.0 | ||
| T/T | 40 | 7 | 33 | 82.5 | 4.60 (1.39-16.69) | .0071 | |
| rs12979860 | Non-C/C | 41 | 18 | 23 | 56.1 | ||
| C/C | 27 | 3 | 24 | 88.9 | 6.10 (1.49-36.64) | .0065 | |
| rs368234815 | Non-TT/TT | 41 | 18 | 23 | 56.1 | ||
| TT/TT | 27 | 3 | 24 | 88.9 | 6.10 (1.49-36.64) | .0065 | |
| rs117648444 | G/G | 53 | 18 | 35 | 66.0 | ||
| G/A | 15 | 3 | 12 | 80.0 | 2.04 (0.46-12.68) | .3609 | |
| Influence of IFNL4 diplotypes on HR in ropeg treatment of PV | |||||||
| IFNL4-P70 alone* | P/N and P/P | 27 | 8 | 19 | 70.4 | Reference | |
| IFNL4-S70 and S70/P70* | N/S and P/S | 21 | 2 | 19 | 90.5 | 3.89 (0.66-42.31) | .1518 |
| No IFNL4* | N/N | 32 | 5 | 27 | 84.4 | 2.24 (0.55-10.16) | .2236 |
| No IFNL4† | N/N | 32 | 5 | 27 | 84.4 | Reference | |
| IFNL4-S70 and S70/P70† | N/S and P/S | 21 | 2 | 19 | 90.5 | 1.74 (0.25-20.11) | .6897 |
| Influence of IFNL4 diplotypes on MR in ropeg treatment of PV | |||||||
| IFNL4-P70 alone* | P/N and P/P | 26 | 15 | 11 | 42.3 | Reference | |
| IFNL4-S70 and S70/P70* | N/S and P/S | 15 | 3 | 12 | 80.0 | 5.23 (1.06 35.87) | .0254 |
| No IFNL4* | N/N | 28 | 3 | 25 | 89.3 | 10.80(2.39-69.97) | 3.91E-04 |
| no IFNL4† | N/N | 28 | 3 | 25 | 89.3 | Reference | |
| IFNL4-S70 and S70/P70† | N/S and P/S | 15 | 3 | 12 | 80.0 | 0.49 (0.06-4.20) | .6474 |
| Variant/diplotype . | Genotype/diplotype status . | n . | NR, n . | R, n . | % R . | OR (95% CI) . | P . |
|---|---|---|---|---|---|---|---|
| Influence of IFNL4 individual variants on HR in ropeg treatment of PV | |||||||
| rs8099917 | Non-TT | 29 | 8 | 21 | 72.4 | ||
| TT | 49 | 7 | 42 | 85.7 | 2.26 (0.62-8.45) | .2336 | |
| rs12979860 | Non-CC | 48 | 10 | 38 | 79.2 | ||
| CC | 31 | 5 | 26 | 83.9 | 1.36 (0.37-5.70) | .7710 | |
| rs368234815 | Non-TT | 48 | 10 | 38 | 79.2 | ||
| TT | 31 | 5 | 26 | 83.9 | 1.36 (0.37-5.70) | .7710 | |
| rs117648444 | GG | 58 | 13 | 45 | 77.6 | ||
| GA | 21 | 2 | 19 | 90.5 | 2.71 (0.53-27.11) | .3303 | |
| Influence of IFNL4 individual variants on MR in ropeg treatment of PV | |||||||
| rs8099917 | Non-T/T | 28 | 14 | 14 | 50.0 | ||
| T/T | 40 | 7 | 33 | 82.5 | 4.60 (1.39-16.69) | .0071 | |
| rs12979860 | Non-C/C | 41 | 18 | 23 | 56.1 | ||
| C/C | 27 | 3 | 24 | 88.9 | 6.10 (1.49-36.64) | .0065 | |
| rs368234815 | Non-TT/TT | 41 | 18 | 23 | 56.1 | ||
| TT/TT | 27 | 3 | 24 | 88.9 | 6.10 (1.49-36.64) | .0065 | |
| rs117648444 | G/G | 53 | 18 | 35 | 66.0 | ||
| G/A | 15 | 3 | 12 | 80.0 | 2.04 (0.46-12.68) | .3609 | |
| Influence of IFNL4 diplotypes on HR in ropeg treatment of PV | |||||||
| IFNL4-P70 alone* | P/N and P/P | 27 | 8 | 19 | 70.4 | Reference | |
| IFNL4-S70 and S70/P70* | N/S and P/S | 21 | 2 | 19 | 90.5 | 3.89 (0.66-42.31) | .1518 |
| No IFNL4* | N/N | 32 | 5 | 27 | 84.4 | 2.24 (0.55-10.16) | .2236 |
| No IFNL4† | N/N | 32 | 5 | 27 | 84.4 | Reference | |
| IFNL4-S70 and S70/P70† | N/S and P/S | 21 | 2 | 19 | 90.5 | 1.74 (0.25-20.11) | .6897 |
| Influence of IFNL4 diplotypes on MR in ropeg treatment of PV | |||||||
| IFNL4-P70 alone* | P/N and P/P | 26 | 15 | 11 | 42.3 | Reference | |
| IFNL4-S70 and S70/P70* | N/S and P/S | 15 | 3 | 12 | 80.0 | 5.23 (1.06 35.87) | .0254 |
| No IFNL4* | N/N | 28 | 3 | 25 | 89.3 | 10.80(2.39-69.97) | 3.91E-04 |
| no IFNL4† | N/N | 28 | 3 | 25 | 89.3 | Reference | |
| IFNL4-S70 and S70/P70† | N/S and P/S | 15 | 3 | 12 | 80.0 | 0.49 (0.06-4.20) | .6474 |
P values by Fisher’s exact test.
N, no IFNL4; NR, nonresponder; P, IFNL4-P70; R, responder; S, IFNL4-S70.
Contribution of IFNL4-S70 (IFNL4-S70 alone + IFNL4-S70/P70) and no-IFNL4, compared with that of IFNL4-P70 alone.
Contribution of IFNL4-S70 (IFNL4-S70 alone + IFNL4-S70/P70), compared with that of no-IFNL4.
Because the strength of association increased with treatment duration (Figure 1C), we performed detailed analyses at a 36-month follow-up, including the noncoding tagSNPs (rs8099917 and rs12979860) and the coding diplotype variants (rs368234815 and rs117648444). First, we studied the association of these IFNL4 variants individually. HR was not significantly influenced by the variants tested (Table 1); however, we observed a significant association of IFNL4 variants with MR. Specifically, rs117648444 alone showed no association, whereas rs368234815 and rs12979860 individually exerted a strong effect on MR, the 2 variants being in complete linkage disequilibrium in our cohort. Furthermore, rs8099917 tagged the causative IFNL4 variants to a lesser extent (Table 1), similar to what has been reported in other studies.11
Next, we evaluated the rs368234815/rs117648444 diplotype for association with HR and MR. Of the 9 existing diplotype combinations derived from phased genotypes, in accordance with described population allele frequencies (supplemental Table 2), only 5 were observed in our cohort (supplemental Table 3). These diploid combinations were further grouped into 3 main functional categories: patients who did not produce any functional IFNL4 (no IFNL4), producers of the impaired IFNL4-S70, and producers of the fully functional IFNL4-P70. Only a small percentage of patients (<6% in HR and MR categories; supplemental Table 3) carried both the S70 and P70 alleles. Analyses based on IFNL4 functionality did not reveal any associations with HR. However, for MR a strong difference in response rates between the patients with no IFNL4 (89.3% responders) and those with IFNL4-P70 (43.3% responders) was observed (P = 3.91 × 10−4; OR, 10.80; 95% CI, 2.39-69.97; Table 1; Figure 1D). Notably, the effect of the impaired variant IFNL4-S70 resembled that of no IFNL4 (Figure 1D). This finding was also reflected in the quantitative response profiles based on JAK2V617F burden, which allowed for stratification of patients by MR according to IFNL4 diploid functional status (Figure 1E; supplemental Figure 3).
To validate the association of the IFNL4 diplotype with MR in an independent cohort of patients with PV, we repeated the analyses of quantitative trait association of MR data in conjunction with phased diplotypes in 27 patients enrolled in the PEGINVERA9 prospective clinical trial. Despite the modest size of this replication cohort, we observed a consistent trend toward improved MRs after ropeg treatment in patients lacking functional IFNL4 that reached borderline statistical significance at the 24-month follow-up (P = .068; supplemental Figure 4).
Germline IFNL4 variants have also been demonstrated to affect therapy-independent, spontaneous hepatitis C viral clearance.23,24 In our PV discovery cohort (n = 122), we did not observe IFNL4 diplotype-dependent differences in JAKV617F burden levels before trial enrollment (supplemental Figure 5), implying that IFN-α treatment has a strong mechanistic role in the effect of IFNL4 on MR.
Our results demonstrate a robust association of IFNL4 diplotype status with MR but not with HR, which may reflect a direct effect of IFN-α on the JAK2V617F burden. More than one-third of patients in our study cohort carried at least 1 allele coding for the fully functional IFNL4-P70 protein variant (supplemental Table 3). Because deep MRs are indispensable for curative therapy, it is important to investigate whether increased treatment duration can overcome the dampening effect of functional IFNL4 on MR. Longitudinal monitoring of the JAK2V617F burden in conjunction with determination of the phased IFNL4 diplotype status, rather than typing tagSNPs, may allow for optimization of patient management during ropeg treatment. Functional studies of this genetic predisposition may provide insight into the interplay between intrinsic expression of type III IFNs and IFN-α therapy.
Original data will be shared upon e-mail request to the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the PROUD-PV study group for collection of clinical data; all the members of the Kralovics Laboratory for discussion and feedback; the Biomedical Sequencing Facility, in particular Donat Alpar and Thomas Winkler-Penz, for assistance and advice on next-generation sequencing; Vesna Krajina for help with data visualization; and all patients enrolled in the PROUD-PV and CONTINUATION-PV clinical trials.
This work was supported by Austrian Science Fund Projects F4702-B20 and P29018-B30 (R.K.). Data generation in part, as well as collection of clinical data used in this study, were funded by AOP Orphan Pharmaceuticals AG.
Authorship
Contribution: R.J. designed and performed experiments and analyzed the data; H.G. contributed to the clinical study design, collection of clinical data, and preparation of the manuscript; E.F., E.B., J.D.M.F., J.W., B.G., and M.S. performed experiments and clinical sampling; F.S. contributed to data analysis and visualization; M.Z., K.K., and C.K. contributed to study coordination, clinical data management, and preparation of the manuscript; R.K. designed experiments and oversaw analyses; and R.J. and R.K. designed the genetic study, performed data interpretation, and wrote the paper.
Conflict-of-interest disclosure: R.K. has received honoraria from and served on the advisory board of AOP Orphan Pharmaceuticals AG; has received honoraria from Pharma Essentia; and has equity ownership in MyeloPro Diagnostics and Research GmbH. H.G. has been a consultant to and received honoraria and research funding from AOP Orphan Pharmaceuticals AG; has received honoraria from Novartis, Celgene, and Janssen-Cilag; has been a consultant to Roche, MyeloPro Diagnostics and Research GmbH; and has received personal fees from PharmaEssentia. M.Z., K.K., and C.K. are employed by AOP Orphan Pharmaceuticals AG. The remaining authors declare no competing financial interests.
Correspondence: Robert Kralovics, Department of Laboratory Medicine, Medical University of Vienna, Vienna, Austria; e-mail: robert.kralovics@meduniwien.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal