Key Points
The immune responses to PF4 and to the spike protein are independent of one another.
Antibodies from patients with VITT and thrombosis do not cross-react with the spike protein.
Abstract
Vaccine-induced immune thrombotic thrombocytopenia (VITT) is a severe adverse effect of ChAdOx1 nCoV-19 COVID-19 vaccine (Vaxzevria) and Janssen Ad26.COV2.S COVID-19 vaccine, and it is associated with unusual thrombosis. VITT is caused by anti-platelet factor 4 (PF4) antibodies activating platelets through their FcγRIIa receptors. Antibodies that activate platelets through FcγRIIa receptors have also been identified in patients with COVID-19. These findings raise concern that vaccination-induced antibodies against anti-SARS-CoV-2 spike protein cause thrombosis by cross-reacting with PF4. Immunogenic epitopes of PF4 and SARS-CoV-2 spike protein were compared using in silico prediction tools and 3D modeling. The SARS-CoV-2 spike protein and PF4 share at least 1 similar epitope. Reactivity of purified anti-PF4 antibodies from patients with VITT was tested against recombinant SARS-CoV-2 spike protein. However, none of the affinity-purified anti-PF4 antibodies from 14 patients with VITT cross-reacted with SARS-CoV-2 spike protein. Sera from 222 polymerase chain reaction–confirmed patients with COVID-19 from 5 European centers were tested by PF4-heparin enzyme-linked immunosorbent assays and PF4-dependent platelet activation assays. We found anti-PF4 antibodies in sera from 19 (8.6%) of 222 patients with COVID-19. However, only 4 showed weak to moderate platelet activation in the presence of PF4, and none of those patients developed thrombotic complications. Among 10 (4.5%) of 222 patients who had COVID-19 with thrombosis, none showed PF4-dependent platelet-activating antibodies. In conclusion, antibodies against PF4 induced by vaccination do not cross-react with the SARS-CoV-2 spike protein, indicating that the intended vaccine-induced immune response against SARS-CoV-2 spike protein is not the trigger of VITT. PF4-reactive antibodies found in patients with COVID-19 in this study were not associated with thrombotic complications.
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a single-stranded RNA virus encoding 16 nonstructural proteins (1-16), 8 accessory proteins (ORF3a, 6, 7a, 7b, 8, 9b, 9c, and 10), and 4 structural proteins known as S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins.1 The spike glycoprotein is responsible for recognition of host cell membrane receptors ACE2 and TMPRSS2 and for mediating fusion with the host cell membrane.2
The European Medical Agency has approved 4 vaccines3 for prevention of symptomatic COVID-19. Two of the vaccines are messenger RNA (mRNA)–based vaccines encoding the spike protein antigen of SARS-CoV-2 encapsulated in lipid nanoparticles, Comirnaty (BioNTech/Pfizer) and COVID-19 mRNA-1273 vaccine (Moderna).
A third vaccine is a recombinant chimpanzee adenoviral vector (ChAdOx1-S) encoding the spike glycoprotein of SARS-CoV-2, ChAdOx1 nCoV-19 COVID-19 vaccine (Vaxzevria; AstraZeneca).
The fourth is a recombinant adenovirus type 26 vector (Ad26.COV2.S) encoding the SARS-CoV-2 spike glycoprotein, the Janssen COVID-19 Vaccine. In Germany since March 2021, about 100 patients4 with venous thromboses at unusual sites (cerebral venous sinus thrombosis and splanchnic vein thrombosis) in combination with moderate to severe thrombocytopenia were observed in individuals ∼5 to 30 days after vaccination with the ChAdOx1 nCoV-19 COVID-19 vaccine.5-7 Similar complications have also been reported after vaccination with the Ad26.COV2.S Janssen COVID-19 vaccine,8,9 known as vaccine-induced immune thrombotic thrombocytopenia (VITT).10 We have identified immunoglobulin G (IgG) class platelet-activating antibodies directed against the cationic platelet chemokine platelet factor 4 (PF4; CXCL4), as the underlying cause of VITT.5
Thromboembolic complications are a major disease burden in hospitalized patients with COVID-19, even in patients without severe respiratory disease. Sometimes thrombosis in patients with COVID-19 also occurs at unusual locations such as in cerebral veins.11-13 However, the overall presentations of the patients with COVID-19 or VITT are quite different. VITT patients often show laboratory signs of disseminated intravascular coagulation with severe thrombocytopenia and were otherwise well before the abrupt onset of thrombosis. In contrast, patients with COVID-19 show disseminated intravascular coagulation typically only with severe disease or as a complication of extracorporeal circulatory support.14
A further similarity between patients with COVID-19 or VITT is IgG-mediated platelet activation via platelet FcγIIa receptors. This has been shown for patients with VITT5 and also by 2 recent studies of patients with COVID-19.15,16 Furthermore, sera from patients with VITT usually react strongly in PF4-heparin enzyme-linked immunosorbent assays (ELISAs), a finding also seen in occasional patients with COVID-19.17 But in contrast to the strong platelet-activating anti-PF4 antibodies from patients with VITT, COVID-19 sera with anti-PF4 antibodies usually are not platelet activating.17
This overlapping clinical picture of unusual thrombotic complications, antibody-induced FcγIIa receptor–dependent platelet activation, and occasional reports of anti-PF4 antibodies in patients with COVID-19, raises the question of whether the immune response against the spike protein induced by vaccination could induce antibodies that cross-react with immunogenic epitopes shared between spike protein and PF4. Accordingly, the overall aim of this study was to determine whether platelet-activating anti-PF4 antibodies in patients with VITT cross-react with the spike protein or whether the anti-PF4 and anti-spike immune responses are distinct. We addressed this by first determining (by using structure analyses in silico) whether there are shared immunogenic epitopes between SARS-CoV-2 spike protein and PF4; second, we used classic immunohematology techniques of immunoadsorption to assess for cross-reactivity of anti-PF4 antibodies against the spike protein obtained from patients with VITT. Our study also examined the presence of VITT-like anti-PF4 antibodies in patients with COVID-19 and their association with thrombosis.
Materials and methods
Identification of immunogenic epitopes and homologies of human PF4 and SARS-CoV-2 spike protein and comparative analysis of their 3D structures
The protein sequence for human PF4 (CXCL4) was retrieved from the ENSEMBL gene database (ENSG00000163737).18 Similarly, the protein sequence of the SARS-CoV-2 spike protein (1273 amino acids) was retrieved from publicly available databases (National Center for Biotechnology Information (NCBI): Gene ID 43740568).19 By using the online prediction tool from the University of Madrid (Madrid, Spain) (http://imed.med.ucm.es/Tools/antigenic.pl),20 we identified potential immunogenic peptide sequences (epitopes) in both protein sequences. We used the SIM Alignment online tool21 and the MacMYPOL program,22 together with the files 6vxx.pbd, 4r9w.pbd, and 4hsv.pbd available from the Protein Data Bank database23 to compare the epitopes on the published structures of these 3 proteins.
Sera were obtained from 24 patients with VITT, defined as patients presenting with thrombocytopenia and thromboembolic events ∼5 to 30 days after vaccination with ChAdOx1 nCoV-19 vaccine with positive PF4-heparin ELISA results and PF4-dependent platelet-activating antibodies tested in the Greifswald Laboratory. The SARS-CoV2 spike ectodomain amino acids 17-1213 and the receptor binding domain (RBD) SD1 amino acids 319-519 (based on QHD43416)24 were cloned and expressed in the human cell line Expi293 (Thermo Fisher Scientific, Langenselbold, Germany). For details, see the supplemental Methods, available online on the Blood Web site.
Testing for PF4-heparin–reactive and platelet-activating IgG antibodies
For screening all sera from the patients with COVID-19 or VITT, we used an IgG-specific anti-PF4-heparin ELISA, and antibody binding was measured by using a secondary anti-human IgG antibody, as described.25 PF4 derived from platelets and recombinant PF4 were obtained from Chromatec (Greifswald, Germany). Optical density (OD) results of <0.5 units were considered negative, ≥0.5 to <1.0 were weak-positive, and OD ≥1.0 were strong-positive.
We performed platelet activation assays by using purified, washed platelets from healthy volunteers (as described),5 by using patient sera, or by using the respective purified anti-PF4 IgG fractions (supplemental Methods) with and without addition of PF4 (10 µg/mL) (Chromatec). Unfractionated heparin (100 IU/mL, final concentration) was added to evaluate inhibition of antibody- and PF4-dependent platelet activation. Platelet activation was judged positive if at least 2 of 3 donor cells aggregated within 30 minutes.26,27
Binding studies of affinity-purified anti-PF4 IgG to SARS-CoV-2 S1 domain, receptor-binding domain, full-length spike protein, PF4, and PF4-heparin complexes
We identified sera that tested positive for anti-PF4-heparin antibodies from 2 patient groups: patients with COVID-19 (only a minority tested positive) and patients with VITT (all tested positive). These sera were assessed for reactivity against spike protein antigens using the following targets: SARS-CoV-2 full-length spike protein and the RDB-SD1 (both assessed using in-house ELISAs), and a commercially available CoV-2 ELISA (recombinant S1 domain; EI 2606-9620 G; EUROIMMUN Medizinische Labordiagnostika AG, Lübeck, Germany). Anti-PF4 and anti-PF4-heparin affinity-purified IgG fractions from 14 patients with VITT (all with documented thromboembolic events) were used in a 1:20 dilution (detailed description is provided in the supplemental Methods).
Cohorts of patients with COVID-19
A total of 222 patients with COVID-19 were enrolled from 5 prospective registries from University Medical Centers in Munich (CORKUM) (World Health Organization [WHO] trial ID: DRKS00021225), Freiburg (WHO trial ID: DRKS00021206), Tübingen (approval by the local ethics committee, trial ID: 240/2018BO2), Greifswald (DRKS-ID: DRKS00023770), and Bari, Italy (approval by the local ethics committee, trial ID: NP 4463).Enrolled patients were between the age of 4 months and 88 years with at least 1 mL available serum or citrate anticoagulated plasma and positive polymerase chain reaction testing of SARS-CoV-2 in nasopharyngeal swabs. Registries began recruiting patients at varying start dates ranging from February 2020 to October 2020. Patient characteristics are summarized in Table 1; registries are described in detail in supplemental Methods; supplemental Tables 1 and 2.
Patient characteristics
| Characteristic . | Patients with COVID-19 . | |
|---|---|---|
| Without thrombosis . | With thrombosis* . | |
| No. of patients | 212 (100) | 10 (100) |
| Sex | ||
| Female | 93 (43.8) | 4 (40.0) |
| Male | 119 (56.2) | 6 (60.0) |
| Age, y | ||
| Median (range) | 55 (0.4-88) | 55 (23-84) |
| <60 | 146 (68.9) | 6 (60.0) |
| ≥60 | 66 (31.1) | 4 (40.0) |
| Outpatient care | 61 (28.8) | 0 |
| Hospitalization | 151 (71.2) | 10 (100) |
| General ward (% of all patients) | 122 (57.5) | 8 (80.0) |
| Intensive care unit (% of all patients) | 29 (13.7) | 2 (20.0) |
| WHO COVID-19 score | ||
| 1-3 | 87 (41.0) | 3 (30.0) |
| 4-5 | 105 (49.5) | 5 (50.0) |
| 6-9 | 18 (8.5) | 2 (20.0) |
| 10 | 2 (0.95) | 0 |
| Interval from symptoms to blood drawing, d | ||
| 0-10 | 115 (54.2) | 4 (40.0) |
| 11-20 | 55 (25.9) | 5 (50.0) |
| 21-50 | 37 (17.5) | 1 (10.0) |
| >50 | 5 (2.4) | |
| Mean platelet count at time of blood drawing × 109/L (range) | 239 (24-769) | 223 (82-364) |
| No. of patients with platelet count (× 109/L) | ||
| >150 | 178 (83.9) | 7 (70.0) |
| >100-150 | 24 (11.3) | 2 (20.0) |
| 50 to ≤100 | 7 (3.3) | 1 (10.0) |
| <50 | 1 (0.47) | 0 |
| Missing data | 2 (0.94) | |
| Heparin treatment (≥5 d) before blood drawing | 32 (15.0) | 3 (30.0) |
| Missing data | 2 (0.94) | 0 |
| PF4-heparin ELISA OD | ||
| <0.5 | 194 (91.5) | 9 (90.0) |
| ≥0.5 to <1.0 | 13 (6.1) | 0 |
| ≥1.0 | 5 (2.4) | 1 (10.0) |
| Heparin-dependent platelet activation, (sera with PF4-heparin ELISA OD ≥0.5) | ||
| Negative | 18 (8.5) | 10† |
| Positive | 0 | 0 |
| PF4-dependent platelet activation (sera with PF4-heparin ELISA OD ≥0.5) | ||
| Negative | 14 (6.6) | 10† |
| Positive | 4 (1.9) | 0 |
| Outcome | ||
| Survived | 206 (97.2) | 10 (100) |
| In-hospital deaths | 6 (2.8) | 0 |
| Characteristic . | Patients with COVID-19 . | |
|---|---|---|
| Without thrombosis . | With thrombosis* . | |
| No. of patients | 212 (100) | 10 (100) |
| Sex | ||
| Female | 93 (43.8) | 4 (40.0) |
| Male | 119 (56.2) | 6 (60.0) |
| Age, y | ||
| Median (range) | 55 (0.4-88) | 55 (23-84) |
| <60 | 146 (68.9) | 6 (60.0) |
| ≥60 | 66 (31.1) | 4 (40.0) |
| Outpatient care | 61 (28.8) | 0 |
| Hospitalization | 151 (71.2) | 10 (100) |
| General ward (% of all patients) | 122 (57.5) | 8 (80.0) |
| Intensive care unit (% of all patients) | 29 (13.7) | 2 (20.0) |
| WHO COVID-19 score | ||
| 1-3 | 87 (41.0) | 3 (30.0) |
| 4-5 | 105 (49.5) | 5 (50.0) |
| 6-9 | 18 (8.5) | 2 (20.0) |
| 10 | 2 (0.95) | 0 |
| Interval from symptoms to blood drawing, d | ||
| 0-10 | 115 (54.2) | 4 (40.0) |
| 11-20 | 55 (25.9) | 5 (50.0) |
| 21-50 | 37 (17.5) | 1 (10.0) |
| >50 | 5 (2.4) | |
| Mean platelet count at time of blood drawing × 109/L (range) | 239 (24-769) | 223 (82-364) |
| No. of patients with platelet count (× 109/L) | ||
| >150 | 178 (83.9) | 7 (70.0) |
| >100-150 | 24 (11.3) | 2 (20.0) |
| 50 to ≤100 | 7 (3.3) | 1 (10.0) |
| <50 | 1 (0.47) | 0 |
| Missing data | 2 (0.94) | |
| Heparin treatment (≥5 d) before blood drawing | 32 (15.0) | 3 (30.0) |
| Missing data | 2 (0.94) | 0 |
| PF4-heparin ELISA OD | ||
| <0.5 | 194 (91.5) | 9 (90.0) |
| ≥0.5 to <1.0 | 13 (6.1) | 0 |
| ≥1.0 | 5 (2.4) | 1 (10.0) |
| Heparin-dependent platelet activation, (sera with PF4-heparin ELISA OD ≥0.5) | ||
| Negative | 18 (8.5) | 10† |
| Positive | 0 | 0 |
| PF4-dependent platelet activation (sera with PF4-heparin ELISA OD ≥0.5) | ||
| Negative | 14 (6.6) | 10† |
| Positive | 4 (1.9) | 0 |
| Outcome | ||
| Survived | 206 (97.2) | 10 (100) |
| In-hospital deaths | 6 (2.8) | 0 |
All data are n (%), unless otherwise indicated. Data were collected from 5 university hospitals: Munich, n = 55; Freiburg, n = 42; Tübingen, n = 32; Greifswald, n = 32; Bari, n = 61.
Thrombosis localization: pulmonary embolism, 6; stroke, 1; portal vein, 2; unknown, 1.
All sera from patients with thrombosis were tested independently of the PF4-heparin ELISA result.
Ethics
All studies of patients with COVID 19 have been approved by the local institutional review board or an independent ethics committee. All patients (or their representatives) provided written informed consent. The use of whole blood and washed platelets from healthy adult individuals and the use of blood from patients with VITT was approved by the Ethics Committee of the University Medicine Greifswald. All volunteers gave written informed consent in accordance with the Declaration of Helsinki. All experiments were carried out in accordance with the approved guidelines.
Results
Similarities between human PF4 and the SARS-CoV-2 spike protein structures
PF4 and the SARS-CoV-2 spike protein show sequence homologies (supplemental Table 1A-B). The spike protein (6vxx.pbd: 323-335) is very similar to 2 consecutive epitopes within PF4 (6-21 and 23-43), although the spike epitope resembles a planar configuration and the PF4 structure is more of a pleated sheet (supplemental Figure 1A). This motif in PF4 is involved in binding heparin (supplemental Figure 1B-C).28 The identical structure is expressed by PF4 variant 1 (details are provided in supplemental Results).
Cohort of patients with VITT
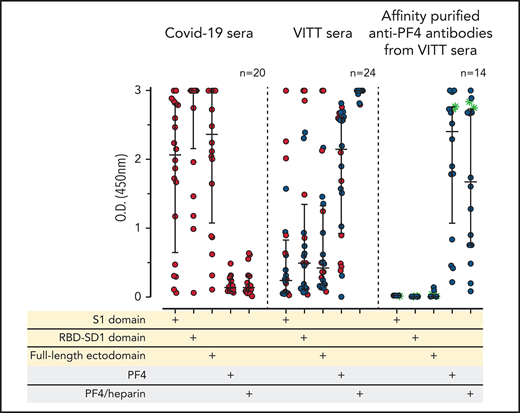
We evaluated sera from 24 patients with confirmed VITT that developed after the first dose of the ChAdOx1 nCoV-19 COVID-19 vaccine. As expected in the context of an early primary immune response, the sera of these (recently vaccinated) 24 patients with VITT contained mostly IgG weakly to moderately binding to the S1 sequence and the RBD sequence of the spike protein, with somewhat higher levels of ELISA reactivity using the full-length spike protein. In contrast, all VITT sera showed binding to PF4 and PF4-heparin complexes, most with high reactivity (OD >2.5; Figure 1A). Because our PF4 preparation contains 1.6% to 2% PF4 variant 1 (as determined by proteome analysis), we also tested 9 of the VITT sera with recombinant PF4 (which lacks PF4 variant1), which showed the same results (supplemental Figure 2). This excludes reactivity caused by binding only to PF4 variant 1. The slightly stronger signal with PF4-heparin complexes is most likely explained by a higher amount of PF4 being available on the plates when they are coated with PF4-heparin complexes instead of single PF4 molecules. The variable reactivity of VITT sera with PF4 alone is probably caused by conformational alteration of PF4 upon coating to the plastic surface, which affects binding of a subset of VITT antibodies. This is consistent with the known variable sensitivities of anti-PF4-polyanion assays for VITT-related anti-PF4 antibodies, which presumably are related to differences in test characteristics, including how the antigen is coated.29-31
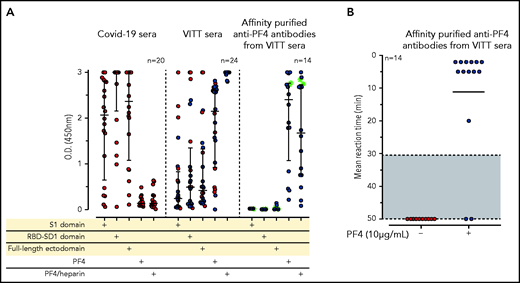
IgG antibodies in patients with COVID-19 or VITT against anti-SARS-CoV-2 spike protein and PF4. (A) Individual OD results of sera tested by ELISA. Error bars are medians with interquartile ranges (indicated by red lines). Graph shows sera of patients with COVID-19 (n = 20), patients with VITT (n = 24), and PF4-affinity-purified (blue stars) IgG or PF4-heparin affinity-purified (^) IgG from VITT sera (n = 14). The 14 sera used for affinity purification of anti-PF4 IgG are indicated by green filled circles. All sera and the respective affinity-purified anti-PF4 IgG fractions were tested against SARS-CoV-2 S1 domain, RBD-SD1 domain, spike full-length ectodomain, PF4, and PF4-heparin complexes. Sera of patients with COVID-19 reacted with the spike protein and its S1 and RBD domains but not with PF4 or PF4-heparin complexes. VITT sera reacted with spike protein epitopes and PF4, but reactions were strongest with PF4-heparin complexes, whereas the affinity-purified anti-PF4 antibody fraction reacted with PF4 and PF4-heparin complexes but not with the spike protein or its S1 and RBD-SD1 domains. All negative controls (n = 15) gave negative results (supplemental Figure 3). The positive controls in the experiment with affinity-purified antibody for binding of antibodies to the S1 domain, the RBD domain, and the spike protein were positive (data not shown). (B) As a control to show that the affinity-purified anti-PF4 antibodies could still activate platelets, we incubated 75 µL washed platelets in Tyrodes buffer with PF4 (10 µg/mL) and added 10 µL of the affinity-purified antibodies, which reacted in the same manner as the original serum. Results of 14 affinity-purified antibody fractions are shown. Twelve showed strong platelet activation in the presence of PF4. Two antibody fractions still reacted positively by PF4-heparin ELISA but no longer activated platelets, most likely as a result of an antibody yield that was too low after affinity purification.
IgG antibodies in patients with COVID-19 or VITT against anti-SARS-CoV-2 spike protein and PF4. (A) Individual OD results of sera tested by ELISA. Error bars are medians with interquartile ranges (indicated by red lines). Graph shows sera of patients with COVID-19 (n = 20), patients with VITT (n = 24), and PF4-affinity-purified (blue stars) IgG or PF4-heparin affinity-purified (^) IgG from VITT sera (n = 14). The 14 sera used for affinity purification of anti-PF4 IgG are indicated by green filled circles. All sera and the respective affinity-purified anti-PF4 IgG fractions were tested against SARS-CoV-2 S1 domain, RBD-SD1 domain, spike full-length ectodomain, PF4, and PF4-heparin complexes. Sera of patients with COVID-19 reacted with the spike protein and its S1 and RBD domains but not with PF4 or PF4-heparin complexes. VITT sera reacted with spike protein epitopes and PF4, but reactions were strongest with PF4-heparin complexes, whereas the affinity-purified anti-PF4 antibody fraction reacted with PF4 and PF4-heparin complexes but not with the spike protein or its S1 and RBD-SD1 domains. All negative controls (n = 15) gave negative results (supplemental Figure 3). The positive controls in the experiment with affinity-purified antibody for binding of antibodies to the S1 domain, the RBD domain, and the spike protein were positive (data not shown). (B) As a control to show that the affinity-purified anti-PF4 antibodies could still activate platelets, we incubated 75 µL washed platelets in Tyrodes buffer with PF4 (10 µg/mL) and added 10 µL of the affinity-purified antibodies, which reacted in the same manner as the original serum. Results of 14 affinity-purified antibody fractions are shown. Twelve showed strong platelet activation in the presence of PF4. Two antibody fractions still reacted positively by PF4-heparin ELISA but no longer activated platelets, most likely as a result of an antibody yield that was too low after affinity purification.
No serologic cross-reactivity of purified anti-PF4 antibodies from serum from patients with VITT with recombinant SARS-CoV-2 spike protein
To determine whether reactivity of VITT sera to the spike protein and to PF4 was a result of cross-reactivity of the same antibodies binding to both proteins or whether antibodies with different specificities were present, we affinity-purified the anti-PF4 antibodies from sera of 14 patients with VITT (from whom sufficient amounts were available), using both PF4 and PF4-heparin complexes. As shown in Figure 1A (right panel), the affinity-purified antibodies once again bound to PF4 and PF4-heparin complexes, which indicates that affinity purification was successful. The weaker reactivity compared with the original sera results from loss and dilution of antibodies during purification. However, we cannot exclude that by affinity purification, a fraction of PF4-specific antibodies in VITT serum was lost because of failed recognition of the chemically modified biotinylated PF4. This may include antibodies that are cross-reactive with spike protein. Therefore, we also affinity-purified antibodies using complexes of 30% biotinylated PF4, 70% native PF4, and heparin. Affinity-purified antibodies strongly activated platelets in the presence of PF4 (Figure 1B). However, none of the affinity-purified antibodies bound to any of the SARS-CoV-2 spike protein constructs (Figure 1A).
Cohorts of patients with COVID-19
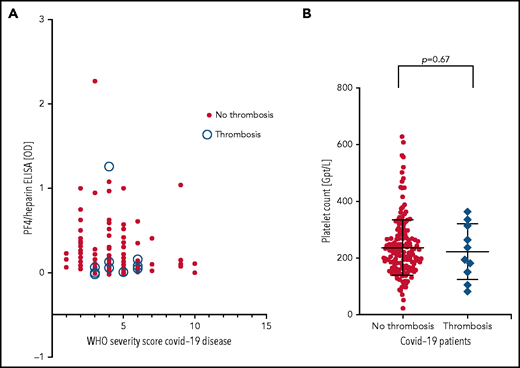
Patients were from 5 medical centers, and sera from a total of 222 patients with COVID-19 (125 males, 97 females; median age, 55 years [range, 4 months to 88 years]) were evaluated in the IgG-specific PF4-heparin ELISA. Nineteen (8.6%) of 222 patients tested positive, with 13 having a result between OD 0.500 and <1.000, and 6 having a result between OD 1.000 and <2.000) (Table 1). There was no correlation between WHO severity score of COVID-19 disease and antibody reactivity by PF4-heparin ELISA (Figure 2A).
PF4-heparin ELISA OD and platelet count in patients with COVID-19. (A) Results of PF4-heparin ELISA OD are given in relation to the WHO Severity Score of COVID-19 disease in 222 patients with COVID-19. ELISA cutoff, 0.5 OD. The 10 patients who developed thrombosis are indicated by open circles. Solid symbols indicate the 212 patients without thrombosis. There was no correlation between WHO severity score of COVID-19 disease and reactivity on the PF4-heparin ELISA. (B) Platelet counts in COVID-19 patients with and without thrombosis.
PF4-heparin ELISA OD and platelet count in patients with COVID-19. (A) Results of PF4-heparin ELISA OD are given in relation to the WHO Severity Score of COVID-19 disease in 222 patients with COVID-19. ELISA cutoff, 0.5 OD. The 10 patients who developed thrombosis are indicated by open circles. Solid symbols indicate the 212 patients without thrombosis. There was no correlation between WHO severity score of COVID-19 disease and reactivity on the PF4-heparin ELISA. (B) Platelet counts in COVID-19 patients with and without thrombosis.
Sera from all 19 patients who tested positive in the anti-PF4-heparin ELISA were tested in the platelet activation assay in the presence of heparin and PF4, respectively, to judge heparin- and PF4-dependent platelet activation. The PF4-enhanced washed platelet activation assay is currently the most sensitive test in our laboratory for detecting VITT platelet-activating antibodies and is more sensitive than the flow cytometric whole blood assay we recently described (data not shown).32 Under reaction conditions previously shown to result in strong serum-induced platelet activation (PF4, 10 µg/mL) (sera were from patients with VITT), we found that 4 of 19 sera showed weak to moderate PF4-dependent platelet activation (lag time median, 15 minutes; range, 10 to 30 minutes). In contrast, none of these sera showed platelet activation in the presence of anti-factor Xa 0.2 U/mL low molecular weight heparin. Thromboembolic complications were reported for 10 (4.5%) of 222 patients (6 with pulmonary embolism, 1 with stroke, 2 with portal vein thrombosis, 1 with thrombosis of unknown localization). Nine of these 10 patients tested negative by PF4-heparin ELISA. Only 1 serum was reactive (OD >1.0), and a pulmonary embolism was reported for this patient. However, none of these 10 sera, including the ELISA-positive patient with pulmonary embolism, induced platelet aggregation in the platelet activation test, regardless of whether heparin or PF4 was added. Moreover, there was no difference in platelet counts in patients with and without thrombosis (Figure 2B).
Discussion
Vaccination against the SARS-CoV-2 spike protein with ChAdOx1 nCov-19 or Janssen Ad26.COV2.S can induce antibodies that cause marked PF4-dependent platelet activation that resulted in thrombocytopenia and unusual thromboses. After SARS-CoV-2 vaccination, most individuals and patients with COVID-19 express antibodies against the spike protein.33-36 Structural analysis of both the spike protein and PF4 indicated potential cross-reactive epitopes. Although the identified linear sequences are similar, the depicted three-dimensional (3D) configurations differ slightly between spike protein and PF4. The relevant structure (323-335) on PF4, however, is a flexible loop, which can also fold on demand (eg, when a high-affinity antibody binds). Moreover, this loop in PF4 has already been shown to change its conformation when heparin binds.28 This raised the concern that vaccination for SARS-CoV-2 might trigger formation of anti-spike protein antibodies that cause VITT by cross-reacting with PF4, resulting in pathogenic PF4-mediated platelet activation.
By using purified recombinant spike protein, purified PF4, and affinity-purified anti-PF4 antibodies from sera obtained from patients with VITT, we found no cross-reactivity between the platelet-activating anti-PF4 antibodies and the spike protein of SARS-CoV-2. Affinity-purified anti-PF4 antibodies from sera of patients with VITT strongly bound in the PF4-heparin ELISA and induced strong PF4-dependent platelet activation. However, the antibodies did not bind to full-length spike protein, the S1 domain, or the RBD-S1 domain. In contrast, most sera tested from patients with COVID-19 contained antibodies that strongly bound to the spike protein, but not to PF4 or PF4-heparin complexes (Figure 1A). This indicates that the immune responses against both proteins, PF4 and spike, are independent of one another.
There are some limitations to our experiments. Affinity purification might preferentially select for a fraction of antibodies in sera from patients with VITT that retains antibody binding to biotinylated PF4 but that lacks cross-reactivity against spike protein if these lost antibodies recognize an epitope covered by biotin on PF4. To address this possibility, we additionally purified antibodies using complexes composed of 30% biotinylated PF4 and 70% native PF4; the resulting affinity-purified antibodies did not bind to any of the spike protein constructs tested. Another limitation is that we could not test spike protein after it has been cleaved by furin or TMPRSS2. Although it is unlikely, we cannot exclude that such cleavage would induce conformational changes allowing binding of anti-PF4 antibodies. A further limitation is that we could not purify anti-spike protein antibodies from patients with COVID-19 (to assess in a reverse fashion for cross-reactivity against PF4) because of limited material for testing.
The reason why patients with VITT produce high-titer platelet-activating anti-PF4 antibodies is currently unknown. PF4 and the related protein PF4 variant 137 have gained major attention in autoimmunity.38-40 Conceivably, predisposition to autoimmunity might lead to disruption of self-tolerance against PF4.
In parallel with these studies, we assessed whether patients with COVID-19 who usually have a strong immune response against the spike protein develop anti-PF4 antibodies similar to those found in patients with VITT, potentially explaining thrombosis associated with COVID-19. However, in a combined analysis of 5 patient cohorts consisting of 222 patients with COVID-19 with variable clinical disease severity (according to the WHO COVID-19 severity score), we found no evidence for an association between anti-PF4-heparin IgG and thromboembolic complications in patients with COVID-19. The frequency of anti-PF4-heparin IgG detectable by ELISA was 8.6%. This number was even lower than that observed in a prospective study in patients in the intensive care unit who did not have COVID-19 (17.2% were anti-PF4-heparin IgG ELISA positive and 5.5% were platelet activation test positive).41 None of the patients with COVID-19 showed heparin-dependent platelet-activating antibodies, and the frequency of PF4-dependent platelet-activating antibodies was only 1.9% (4 of 222). Moreover, the reactivity of sera from these 4 patients with COVID-19 was weak compared with the generally strong reactivity seen with sera from patients with VITT (lag time median, 15 minutes vs <2 to 5 minutes, respectively).
Overall, patients with COVID-19 with and without anti-PF4 antibodies or PF4-dependent platelet-activating antibodies showed similar clinical characteristics. In particular, none of the patients with PF4-dependent platelet-activating antibodies developed thrombosis. In our multicenter cohort of patients with COVID-19, thromboembolic events occurred in 4.5% of patients, and no cerebral vein thrombosis or splanchnic vein thrombosis was reported. Only 1 patient with thrombosis was reactive in the PF4-heparin ELISA, but that patient’s serum did not activate platelets in the presence of either heparin or PF4. This indicates that thrombotic events in patients with COVID-19 are not typically associated with the presence of the same anti-PF4 platelet-activating antibodies identified in vaccinated people who develop VITT. This does not exclude that on rare occasions, patients with COVID-19 could develop prothrombotic PF4-dependent antibodies that activate platelets, but this remains to be established. However, such a phenomenon would be independent of the immune response against the SARS-CoV-2 spike protein, which is highly prevalent in this patient population.
On the basis of our findings taken together, it is unlikely that the intended immune response against the SARS-CoV-2 spike protein itself induces severe VITT by inducing anti-spike protein antibodies cross-reacting with PF4 (or PF4 variant 1). This information is critical for further risk-benefit assessment of the ongoing large vaccination programs. Our study indicates there is no apparent need to change the SARS-CoV-2 spike protein antigen target for the vaccination strategy to curtail the pandemic.
Acknowledgments
The authors thank Sarah Gekeler (University Tübingen), Alexandra Nieters (FREEZE-Biobank [Universitätsklinikum Freiburg und der Medizinischen Fakultät unterstützte Zentrum für Biobanking], University Medical Center Freiburg and Medical Faculty, University Freiburg), and Theresa Winter (University Greifswald) and their teams for excellent logistical and technical support of biobank facilities; Matthias Nauck and Katja Riemann for the Virus-Induced Pneumonia (VIP) Biobank; Uwe Völker for coordinating the VIP study; Nanette Piasta, Manuela Gerber, and Eik Schäfer for recruiting patients; laboratory assistants Ulrike Strobel, Jessica Fuhrmann, Carmen Freier, Ricarda Raschke, Ines Varnig, and Nicole Lembke for managing samples and laboratory testing; Antje Westphal for collecting data and documenting results; and all local investigators, staff, and especially our patients and their families for their participation in the 5 independent patients’ registries.
This work was supported by funders from Munich, Germany (CORKUM): in part by the Federal Ministry of Education and Research (BMBF) NaFoUniMedCOVID19 initiative (01KX2021), Ludwig Maximilians University (LMU) of Munich and the LMU Faculty of Medicine; from Freiburg, Germany: by a grant from the German Center for Infection Research and the Federal Ministry of Education and Research, Germany (8039801926); from Tübingen, Germany: the German Research Foundation (DFG) (Project no. 374031971-TRR 240) and the Ministry of Science, Research and the Arts of the State of Baden-Württemberg (COVID-19 funding); from Greifswald, Germany: by the DFG (Project no. 374031971-TRR 240), by a “Gerhard Domagk” research grant awarded by the University of Greifswald (G.L. and L.S.), by the Friedrich-Loeffler Institut, and by the European Virus Archive Global (EVA-GLOBAL) project funded by a grant from the European Union’s Horizon 2020 research and innovation program (871029); and from Frankfurt am Main, Germany: by grants from DFG (Ma 1876/12-1 and Ma 1876/13-1), from the Wilhelm Sander Foundation (2018.070.1 and 2018.070.2), and from the Corona-Funding Fonds of Goethe University (R.M.). The funders listed above had no role in designing the study, collecting, analyzing, or interpreting the data, or writing the manuscript.
Authorship
Contribution: A.G., K.S., and T.T. conceived the study; A.A. and S.R. produced the recombinant spike protein constructs; J.M., M.M., J.C.H., and O.T.K. developed the COVID-19 Registry of the LMU Klinikum (CORKUM) and organized sample and data transfer; D.D., A.L., and S.R. collected reported cases from the Freiburg cohort and organized sample and data transfer; M.P.G. and K.A.L.M. collected reported cases from the Tübingen cohort and organized sample and data transfer; C.S.S., M.N., and K.H. collected reported cases from the Greifswald cohort and organized sample and data transfer; G.L., A.V., A.F., P.L., and A.S. collected reported cases from the cohort from the University of Bari Aldo Moro, Bari, Italy, and organized sample and data transfer; R.M. provided the structural analysis and comparison of the PF4 and spike protein; R.P. and J.W. performed the IgG affinity purification and in vitro antibody cross-reactivity experiments, analyzed the data, and prepared the figure; A.G., K.S., and T.E.W. analyzed the data; A.G., K.S., J.M., T.E.W., and T.T. wrote the manuscript; and all authors have critically revised and approved the final version of the manuscript.
Conflict-of-interest disclosure: A.G. received grants and non-financial support from Aspen, Boehringer Ingelheim, MSD, Bristol Myers Squibb, Bayer Healthcare, and Instrumentation Laboratory; personal fees from Aspen, MSD, Macopharma, Bristol Myers Squibb, Chromatec, and Instrumentation Laboratory; and non-financial support from Portola, Ergomed, Biokit outside the submitted work. K.S. received lecture fees from Aspen Germany and travel support from the Swedish Orphan Biovitrum. J.M. received research grants from BMBF, DFG, and Deutsche Krebshilfe; received honoraria from the Falk Foundation; and has patents with META-PAC (Plasma-Metabolom Multimarkerpanel für die Diagnose des Pankreaskarzinoms in Risikogruppen). J.C.H. received a grant from LMU. O.T.K. received grants from the Federal Ministry of Education and Research “NaFoUniMedCovid19” (01KX2021) and LMU. M.P.G. received a grant from DFG (374031971-TRR 240), Ministry of Science, Research and the Arts of the State of Baden-Württemberg (COVID-19 funding). K.A.L.M. received a grant from DFG (374031971-TRR 240), Ministry of Science, Research and the Arts of the State of Baden-Württemberg (COVID-19 funding). G.L. holds a patent on SARS-CoV2 vaccine. A.V. received funding from the Käthe-Kluth-Research Group and Else-Kröner-Fresenius-Stiftung. T.T. received grants from DFG during the conduct of the study; personal fees and other from Bristol Myers Squibb, Pfizer, and Chugai Pharma; personal fees from Bayer and Novartis; other from Novo Nordisk and Daichii Sankyo outside the submitted work. The remaining authors declare no competing financial interests.
A complete list of the members of the for the Immune-Response in COVID-19 Vaccination Study Group appears in “Appendix.”
Correspondence: Andreas Greinacher, Universitätsmedizin Greifswald, Institut für Immunologie und Transfusionsmedizin, Abteilung Transfusionsmedizin, Sauerbruchstr, 17487 Greifswald, Germany; e-mail: andreas.greinacher@med.uni-greifswald.de.
Data will be made available to researchers upon reasonable request. For data presented in this article, please contact Andreas Greinacher via e-mail at andreas.greinacher@med.uni-greifswald.de.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Appendix
The members of CORKUM are: Michael von Bergwelt-Baildon, Stefan Kääb, Clemens Scherer, Stefan Endres, Bernd Zwiβler, Martin Reincke, Konstantin Stark, Steffen Massberg, Hans-Christian Stubbe, Paul R. Wratil, Ludwig-Maximilians-Universität München, Munich, Bavaria, Germany Giuseppe De Palma, University of Brescia, provided samples and cared for study patients. Konstanze Aurich, Department of Transfusion Medicine, University Medicine Greifswald, reviewed the results and the manuscript, and organized cohort laboratory investigations. Khalil Eldebuch, Department of Transfusion Medicine, University Medicine Greifswald, helped with testing of the affinity purified antibodies in the functional test. The member of the VIP study group are: Nanette Piasta (Aneasthesiology); Matthias Napp (Emergency Department, Funtional Genomics), Funktionelle Genomforschung Uwe Völker, Manuela Gesell Salazar; Nils Hübner (Hygiene); Matthias Nauck, Katja Riemann, Mario Schattschneider, Christian Schäfer, Theresa Winter (Institut für Klinische Chemie und Laboratoriumsmedizin); Barbara Bröker, Silva Holtfreter, Daniel Mrochen (Department of Immunology); Stephan Felix, Marcus Dörr, Jens Fielitz (Cardiology); Karsten Becker, Kathrin Zimmermann, Jürgen Bohnert (Microbiology); Robert Fleischmann (Neurology); Mladen Tzvetkov, Eik Schäfer (Pharmacology); Dagmar Waltemath (Center for Data Integration); Wolfgang Hoffmann (Trusted Third Party of the University Medicine Greifswald); University Medicine Greifswald, Germany, provided and cared for study patients, collected data.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal