Abstract
Exome and genome sequencing has facilitated the identification of hundreds of genes and other regions that are recurrently mutated in hematologic neoplasms. The data sets from these studies theoretically provide opportunities. Quality differences between data sets can confound secondary analyses. We explore the consequences of these on the conclusions from some recent studies of B-cell lymphomas. We highlight the need for a minimum reporting standard to increase transparency in genomic research.
Introduction
The clinical and translational value of high-throughput sequencing (HTS) to analyze genomes and its use in the study of cancer and other genetic diseases is well established. The role of sequencing in detecting somatic mutations that may be actionable or facilitate assignment of patients to molecular subgroups is becoming standard in a growing array of settings. Projects such as The Cancer Genome Atlas applied whole-exome sequencing (WES) to catalog the common sites of mutation in cohorts of hundreds of patients but prioritized solid tumors, with only a single hematologic malignancy (AML) thoroughly studied in The Cancer Genome Atlas.1,2 Multiple hematologic neoplasms were studied by whole-genome sequencing (WGS) in the International Cancer Genome Consortium (ICGC), namely chronic lymphocytic leukemia,3 diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma (BL), and follicular lymphoma.4 Many WES- or WGS-based studies of these neoplasms and other hematologic cancers have followed.5-9 Using recent publications on BL10-12 and DLBCL13-18 as examples (Figure 1A), we explore how experimental design and methodology can influence the results of genome-wide surveys of cancer genomes. We caution the community that the completeness and accuracy of HTS studies is highly variable and offer some suggestions that could enhance the interpretability of such experiments.
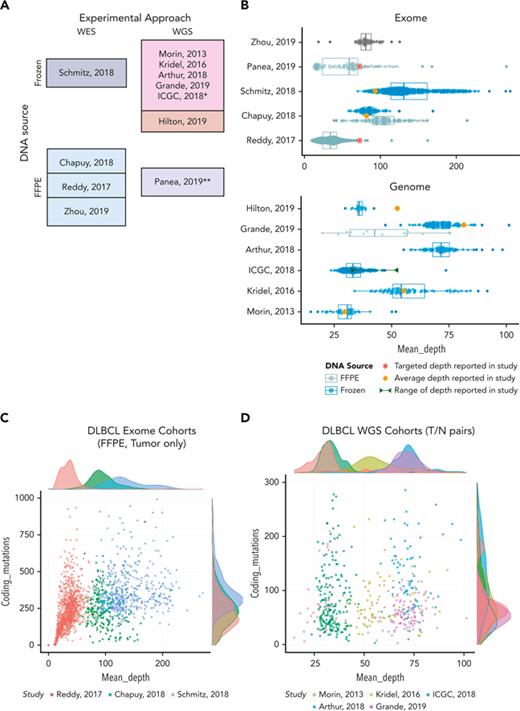
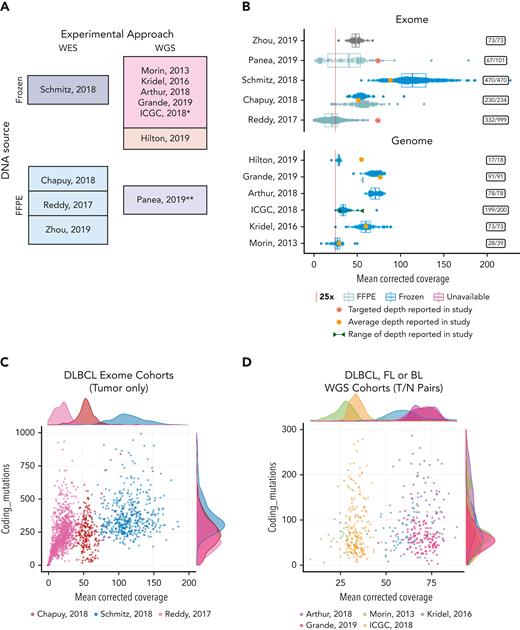
Coverage and mutation burden in samples from selected genomics studies. (A) The assay type, experimental design, and DNA source is shown for each study selected for comparison. WGS studies in the darker shade of pink also performed sequencing on matched normal samples and the remaining studies shown all (or primarily) sequenced only tumors. (B) Box-whisker plot showing the effective nonredundant coverage across the target space or genome (MeanCorrectedCoverage), respectively for the samples from WES (top) or WGS (bottom) studies. Samples from fresh frozen or FFPE tissue are shown separately where that information was available. Individual points showing the coverage of each sample are overlaid. The average depth reported in each study or, when not reported, the targeted coverage is indicated. (C-D) All samples from 8 studies were subjected to the same in-house variant calling pipeline to determine the number of coding variants we could detect. These values are plotted as a function of coverage and shown separately for WES and WGS studies. Insets on the right-hand side show the number of cases in each study with MeanCorrectedCoverage of at least 25×. ∗Data from the ICGC MALY-DE project were used in a series of studies. We use ICGC to refer to all the cases available through EGA. ∗∗Panea et al12 described WGS, but the data deposited in EGA contained both WGS and WES data combined. The original bam files containing both data types were used here, and thus we present them as WES data for comparison. FFPE, formalin-fixed paraffin-embedded.
Coverage and mutation burden in samples from selected genomics studies. (A) The assay type, experimental design, and DNA source is shown for each study selected for comparison. WGS studies in the darker shade of pink also performed sequencing on matched normal samples and the remaining studies shown all (or primarily) sequenced only tumors. (B) Box-whisker plot showing the effective nonredundant coverage across the target space or genome (MeanCorrectedCoverage), respectively for the samples from WES (top) or WGS (bottom) studies. Samples from fresh frozen or FFPE tissue are shown separately where that information was available. Individual points showing the coverage of each sample are overlaid. The average depth reported in each study or, when not reported, the targeted coverage is indicated. (C-D) All samples from 8 studies were subjected to the same in-house variant calling pipeline to determine the number of coding variants we could detect. These values are plotted as a function of coverage and shown separately for WES and WGS studies. Insets on the right-hand side show the number of cases in each study with MeanCorrectedCoverage of at least 25×. ∗Data from the ICGC MALY-DE project were used in a series of studies. We use ICGC to refer to all the cases available through EGA. ∗∗Panea et al12 described WGS, but the data deposited in EGA contained both WGS and WES data combined. The original bam files containing both data types were used here, and thus we present them as WES data for comparison. FFPE, formalin-fixed paraffin-embedded.
Drinking from the data oasis
National Cancer Institute–driven projects implemented defined standards to ensure the generation and release of consistently high-quality data. These standards included requirement for a germ line (reference) control, consistency in pathology review and sample preparation, and quality control criteria at each step of data generation and analysis. Such scrutiny can lead to a significant reduction in the number of samples suitable for genomic studies. Thus, the data from comprehensive “omic” experiments come at an enormous cost, not just in sequencing but in the time involved in annotating and triaging samples. Thankfully, some data sets can be obtained by researchers for secondary analysis, typically from controlled-access repositories. Because of a lack of reporting standards, differences in bioinformatics pipelines and variable degrees of quality control, understanding these data sets generally requires complete reanalysis and careful scrutiny of individual samples, an expensive and time-consuming process likely to be impractical for many. When performing such analyses, some elements that can influence the comparability of data sets were identified.
A search for common ground
Multiple HTS-based surveys of many cancers may use different assays or focus on different patient populations/subtypes, and thus results tend to be complementary while largely in agreement. When there are notable discrepancies in the significantly mutated genes (SMGs) reported or the frequency of mutations therein, it can be difficult for readers to discern the underlying causes. The mutational landscape of DLBCL has been particularly dynamic, with SMGs ranging in number from 32219 to 15013 in studies from a single group. The latter number is closer to that reported in another large WES-based study (98), and yet these lists only share 62 genes.14 In other words, these 2 studies disagree on more genes than they agree on! If we can conclude anything from this, it is that any gene list from a single study is probably incomplete. As part of the scientific process, we can accept that the list of SMGs in any one entity will remain a moving target, migrating with increases in data scale, comprehensiveness, and cohort size.
Among the genes that are consistently reported, comparable mutation frequencies across studies are expected. Although generally true, some striking exceptions exist. For instance, the coding mutations in TP53 in 3 recent studies of BL10-12 are reported to be somewhere between 5% and 50%. In other cases, comparisons can be more difficult to resolve without access to the raw data. For example, H1-4 (HIST1H1E) might be mutated in anywhere from 0 (mutations not reported11), 8%,10 or 42%12 of BLs, and yet the mutation rate of this gene appears much lower in our reanalysis of the same data. Each such situation begs the question: which (if any) is the correct value and what variables, biological or technical, might explain such discrepancies?
Thorny issues
Although discrepant findings may be caused by a combination of patient demographics, clinical setting, experimental methodology, or analytical approach, it is critical to discern biological variation from technical influences. One confounder when comparing studies is whether a source of matched constitutional (or germ line) DNA was included. Blood and buccal sources of DNA can be contaminated with mutations from malignant cells in many of the myeloid cancers, lymphoid leukemias, and other lymphoid cancers such that other tissues are required.20 Whole blood may also include mutations representing clonal hematopoiesis of indeterminate potential.21 In cases of germ line contamination, the standard practice of removing mutations with significant support in the matched sample can lead to the under-reporting of somatic variants. In contrast, in situations where germ line DNA was not considered, we must be careful to avoid mistaking rare or common polymorphisms for somatic mutations. In one such example, a polymorphism in CCNF was reported as SMG in endemic BL, but the variant was not reproduced in subsequent studies that relied on tumor/normal pairs.22 On the other hand, the common practice of applying strict filters to remove common polymorphisms can artificially redact some somatic mutations in the process. Each bioinformatics pipeline has its own unique trade-off of false-positive rate and false-negative rate, but analyses using matched normal reference samples can improve upon both measures. Besides informing on somatic status, a second benefit of paired sequencing is the natural reduction of systematic sequencing artifacts or false-positive variants arising from the incorrect placement of reads from regions with high sequence similarity. KMT2C (MLL3), for example, has been reported as significantly mutated in DLBCL13,19,23,24 but is known to be affected by this issue.25 The shorter fragment lengths of degraded DNA can further reduce the accuracy of read mapping, leading to spurious findings even when a matched normal is sequenced.26
A moving target in an ever-changing landscape
Many hematopathologists and treating physicians will remember the annoyances of competing nomenclatures and appreciate the benefits of the relatively stable diagnostic framework afforded by the modern WHO classification.27,28 Unfortunately, most descriptive terms relating to HTS experiments are only loosely defined. For example, the term “exome” itself is ambiguous because it is dictated by the manufacturers of reagents and does not indicate whether exome enrichment strategies include untranslated regions. Similarly, there are various uses of “coverage” that depend on the computational pipeline used. Because of DNA damage, sequencing error, and coverage evenness varying with sample quality, there is no consistent depth at which the “whole” genome or exome of a sample can be considered adequately sequenced for all applications. Early WGS studies strived for an average coverage around 30×.7 With falling costs and more reliance on FFPE specimens, target depths of 80× or higher are becoming common for WGS.10 This continues to shift because of ongoing evolution of sequencing technologies, which affects throughput, read length and sequencing error profiles, along with our increased understanding of the subclonal diversity of different cancer types. An early step in repurposing and combining HTS data is to determine the relative coverage between samples in each study and, if necessary, remove those having insufficient quality. Within pan-cancer analysis of whole genomes, they used number of reads per cancer chromosome copy as a metric to represent the depth relative to the inferred purity and ploidy of the specimen to compare the relative sampling from tumor cells between experiments.29 To illustrate the case, for our analysis we computed several coverage metrics for each sample using an open-source pipeline (https://tinyurl.com/HTSQC) and compared samples from the selected studies using MeanCorrectedCoverage (supplemental Table 1, available on the Blood website).
Deep sequencing or laying it on thick?
Across the studies evaluated, the achieved coverage genome-wide or in target regions spans a startling range. This is partly a function of the age of the study, but there is also widespread variability within studies. Importantly, the actual usable sequence data and its variation across samples is not conveyed by the common practice of reporting of study-wide averages and is further obfuscated when the authors instead report study-wide “targets” (Figure 1B). Indeed, the per-sample coverage is positively correlated with the number of coding point mutations we can detect, particularly in lower coverage and/or low purity settings (ie, lower values of number of reads per cancer chromosome copy). This indicates that more data would be required to consider such cases adequately sequenced. This relationship is apparent in many samples from a DLBCL study (Figure 1C), but at higher coverages, this correlation diminishes and can saturate because most clonal variants will be detected, as seen in the other studies (Figure 1D). If we directly compare the mutational frequencies in the DLBCLs with high (above 25×) and low (below 25×) MeanCorrectedCoverage, the frequency of mutations affecting known lymphoma genes is strikingly different (supplemental Figure 1) and the difference is statistically significant for many genes (supplemental Figure 2). This illustrates potential effect of variable sequencing depths on the overall conclusions and further highlights the need for consistent, transparent reporting of this information. As the reader may appreciate, using terms such as WES or WGS to describe an experiment is about as useful as assigning a diagnosis of “lymphoma.” As the definitions have been refined in hematology, we must improve our language for describing data from HTS experiments. Many of us would be wary of a clinical study that lacked the obligatory “Table 1,” detailing the patient demographics and clinical characteristics. Shouldn’t genomics researchers be held to a similar standard of transparency?
Recommended information for reporting sample sequencing details
| Attribute . | Attribute type . | Description . |
|---|---|---|
| Unique sample identifier | Identifier | The unique identifier for the sample |
| Component | Sample metadata | Category of sample type referring to experimental design (eg, tumor, normal, cell-free) |
| Tissue preservation | Sample metadata | Category of preservation method for tissue/sample (eg, frozen, FFPE) |
| Sequencing assay type | Assay metadata | Generic name for the assay used to generate the data (eg, WGS, WES) |
| Sequencing platform | Assay metadata | Unambiguous name of sequencing platforms used to generate the data for the sample |
| Target capture regions URL | Assay metadata | Link to a BED format file that specifies the regions targeted for sequencing (when applicable)∗ |
| Aligner | Analysis metadata | Generic name for the workflow/software used to generate alignments (eg, bwa-mem, minimap2) |
| Genomic reference | Analysis metadata | Exact version of the human genome reference used in the alignment of reads |
| Genomic reference URL | Analysis metadata | Link to human genome sequence. URL |
| Average insert size | Quality metric | Average insert size collected from samtools. Integer |
| Average read length | Quality metric | Average read length collected from samtools. Integer |
| MeanCorrectedCoverage | Quality metric | Mean coverage of whole genome or targeted regions, correcting for overlapping regions of reads, collected from Picard. Number |
| Pairs on diff chromosome | Quality metric | Pairs on different chromosomes collected from samtools. Integer |
| Total reads | Quality metric | Total number of reads per sample. Integer |
| Total uniquely mapped | Quality metric | Number of reads that map to genome. Integer |
| Total unmapped reads | Quality metric | Number of reads that did not map to genome. Integer |
| Proportion reads duplicated | Quality metric | Proportion of duplicated reads collected from samtools. Number |
| Proportion reads mapped | Quality metric | Proportion of mapped reads collected from samtools. Number |
| Proportion targets no coverage | Quality metric | Proportion of targets that did not reach 1× coverage over any base. Number∗ |
| Proportion coverage 10× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 10× or greater coverage |
| Proportion coverage 30× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 30× or greater coverage |
| Proportion coverage 100× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 100× or greater coverage∗ |
| Attribute . | Attribute type . | Description . |
|---|---|---|
| Unique sample identifier | Identifier | The unique identifier for the sample |
| Component | Sample metadata | Category of sample type referring to experimental design (eg, tumor, normal, cell-free) |
| Tissue preservation | Sample metadata | Category of preservation method for tissue/sample (eg, frozen, FFPE) |
| Sequencing assay type | Assay metadata | Generic name for the assay used to generate the data (eg, WGS, WES) |
| Sequencing platform | Assay metadata | Unambiguous name of sequencing platforms used to generate the data for the sample |
| Target capture regions URL | Assay metadata | Link to a BED format file that specifies the regions targeted for sequencing (when applicable)∗ |
| Aligner | Analysis metadata | Generic name for the workflow/software used to generate alignments (eg, bwa-mem, minimap2) |
| Genomic reference | Analysis metadata | Exact version of the human genome reference used in the alignment of reads |
| Genomic reference URL | Analysis metadata | Link to human genome sequence. URL |
| Average insert size | Quality metric | Average insert size collected from samtools. Integer |
| Average read length | Quality metric | Average read length collected from samtools. Integer |
| MeanCorrectedCoverage | Quality metric | Mean coverage of whole genome or targeted regions, correcting for overlapping regions of reads, collected from Picard. Number |
| Pairs on diff chromosome | Quality metric | Pairs on different chromosomes collected from samtools. Integer |
| Total reads | Quality metric | Total number of reads per sample. Integer |
| Total uniquely mapped | Quality metric | Number of reads that map to genome. Integer |
| Total unmapped reads | Quality metric | Number of reads that did not map to genome. Integer |
| Proportion reads duplicated | Quality metric | Proportion of duplicated reads collected from samtools. Number |
| Proportion reads mapped | Quality metric | Proportion of mapped reads collected from samtools. Number |
| Proportion targets no coverage | Quality metric | Proportion of targets that did not reach 1× coverage over any base. Number∗ |
| Proportion coverage 10× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 10× or greater coverage |
| Proportion coverage 30× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 30× or greater coverage |
| Proportion coverage 100× | Quality metric | Proportion of all reference bases for WGS or targeted bases that achieves 100× or greater coverage∗ |
All fields are recommended for WGS and targeted sequencing experiments with the exception of those marked with an asterisk, which are required only for targeted sequencing.
Although it would be convenient if every study achieved some “ideal” target depth, this is impractical. Different biological questions can be answered at different sequencing depths. Furthermore, the volume of raw data generated is dictated by biological and budgetary considerations, whereas the amount of usable data is influenced by technical variables. These jointly affect the accuracy of results and may significantly affect mutation frequencies, thereby potentially affecting biological or clinical conclusions. In theory, samples with insufficient sequencing depth or purity should be excluded from analysis, but the exclusion criteria will likely depend on the application. Some material may be fairly rare; therefore, maximizing the value from low-quality samples is critical and the inclusion of samples with suboptimal data may be warranted. We note that in the largest DLBCL WES study, 66% of samples would be eliminated if a very modest threshold of 25× corrected coverage was applied (Figure 1B). It is key to identify technical variables such as this to help identify potential causes of conflicting results.
Out in the open
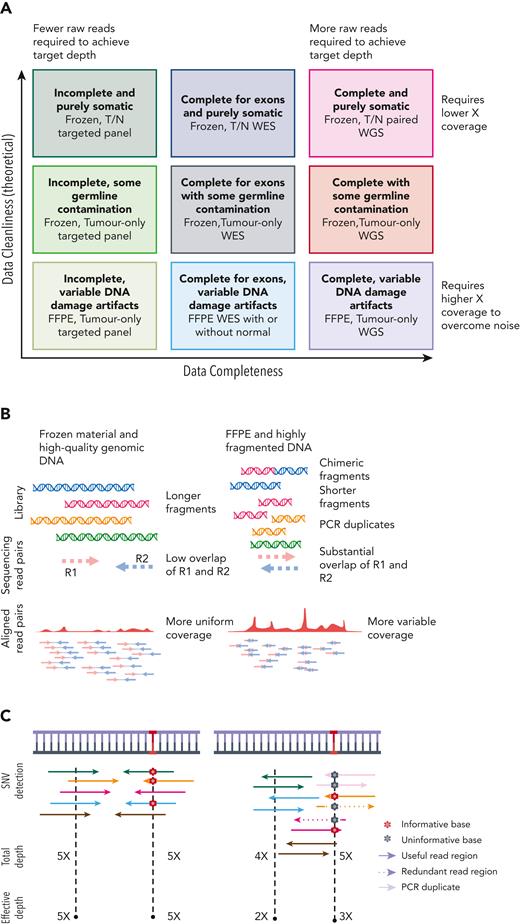
The sequencing coverage (or depth) is alone insufficient as a metric for the discovery potential of an experiment.30 Other features affecting library quality include the fragment length distribution and diversity, which influence the redundancy required to accurately detect somatic mutations (Figure 2A). For technical reasons (Figure 2B), libraries generated from FFPE material require more raw sequencing data to obtain reasonable sensitivity and specificity across the regions of interest. For WGS, variables such as evenness of coverage and proportion of read pairs that span 2 chromosomes can inform on the usefulness of the data, respectively, for copy number and rearrangement detection.30 However, in case of FFPE material, often severely high background of structural variations because of sample degradation makes use of these samples virtually impossible. These factors are partly resolved by reporting the effective coverage or the average coverage after correcting for redundant portions of overlapping reads (Figure 2C).
Experimental variables affecting data quality and accuracy for detecting mutations. (A) Panel-based, WES and WGS are compared in their completeness and requirements for volume of raw data (reads). Each method can be applied to tumors in isolation or with matched germ line DNA also sequenced (T/N pairs). Only common single nucleotide polymorphisms can be recognized from variants detected in unmatched tumors, whereas rare germ line variants and private mutations can be removed if the germ line is sequenced. (B) Frozen tissue is a preferred source of genomic DNA. FFPE-derived DNA has various forms of DNA damage and can be highly fragmented, leading to more overlapping read pairs and more redundant sequence information from each fragment. (C) It has been common to report the average total depth of depth regardless of fragment length or read overlap. Effective (or corrected) depth accounts for the redundant information from overlapping reads and is more consistent with how variant calling algorithms detect mutations.
Experimental variables affecting data quality and accuracy for detecting mutations. (A) Panel-based, WES and WGS are compared in their completeness and requirements for volume of raw data (reads). Each method can be applied to tumors in isolation or with matched germ line DNA also sequenced (T/N pairs). Only common single nucleotide polymorphisms can be recognized from variants detected in unmatched tumors, whereas rare germ line variants and private mutations can be removed if the germ line is sequenced. (B) Frozen tissue is a preferred source of genomic DNA. FFPE-derived DNA has various forms of DNA damage and can be highly fragmented, leading to more overlapping read pairs and more redundant sequence information from each fragment. (C) It has been common to report the average total depth of depth regardless of fragment length or read overlap. Effective (or corrected) depth accounts for the redundant information from overlapping reads and is more consistent with how variant calling algorithms detect mutations.
A path forward
Many standards exist for ensuring consistency in the performance and/or reporting of specific types of experiments, but the proposed HTS standard31 is not easily adopted for research on human subjects and does not consider factors such as DNA quality from archival tissues. In hopes of avoiding the trap of proposing a data standard that would never be adopted, we suggest a minimal set of quality metrics that could enable the objective evaluation and comparison of data completeness and quality. Table 1 shows our proposed standard for reporting WGS and WES experiments, on a per-sample basis, that will facilitate transparent and consistent description of data quality while preserving patient anonymity. Although some of the reporting metrics proposed here can be readily inferred by potential secondary users, this process requires obtaining access to the data, which can be surprisingly arduous. This standard is based directly from the Human Tumor Atlas Network standard for bulk DNA sequencing experiments (https://data.humantumoratlas.org/standard/bulkdnaseq). This set of metrics is equally relevant for genomic studies of other conditions such as clonal hematopoiesis of indeterminate potential and studies seeking rare germ line or de novo variants of potential relevance to disease. The information required from each sample includes a unique sample identifier, preanalytic variables such as sample preservation technique (frozen or FFPE), other sample details such as tumor/normal status, library preparation type, and the sequencing platform. Details of the initial standard analytical steps are also required, including the aligner and the specific genome reference build. The actual quality metrics may vary according to the sequencing technology but include total reads and the number of aligned and nonduplicate reads. Metrics relating to the quality of the library include an estimate of chimeric fragments (approximated from pairs mapping to different chromosomes) and average insert size. In addition to reporting the average coverage of target regions (or genome-wide), we suggest tabulation of the proportion of those regions having at least N-fold coverage, where the authors should report this for N = 30 and at least one higher value of N that represents the desirable coverage for a specific application (eg, 100).32 This standard also requires the mean coverage to be reported after correcting for overlapping regions of reads (MeanCorrectedCoverage) to convey the effective coverage resulting from unique molecules. Importantly, these variables can all be affected by sample quality and are particularly relevant in comparing data from degraded sources of DNA such as FFPE tissues.
Conclusion
When evaluating genomic research, it can be difficult to discern a data oasis from a mirage. We call on the community to adopt a reporting standard that ensures studies contain sufficient details, allowing for adequate scrutiny of their quality and, where feasible, consolidation in meta-analyses. Although there is no shortage of them, widespread adoption of a community standard likely represents the exception rather than the rule. We hope the ASH family of journals will strongly encourage – if not require – adherence to this proposed data reporting standard. A more widespread adoption of MIRAGE or comparable standard across the hematology research community would be collectively beneficial. This would require that authors provide details on data quantity and quality for each sample. Such information is often requested during peer review, but the provided quality control metrics and methods of their collection are inconsistent. Regardless of the nuances of the approach, it must address an unmet need in genomic and germ line research, namely a common language for communicating experimental details to empower stakeholders to assess the value and potential caveats of individual data sets.
Acknowledgments
Some of the data described in this study are available from the European Genome-phenome Archive at the European Bioinformatics Institute (EGAS00001003778, EGAS00001002606, EGAS00001003719) and can be accessed through a request as detailed on the EGA website. The data were used in a form agreed by the User Institution with DAC for Dave Lab, Duke University. The results published here are in whole or in part based upon data generated by the Cancer Genome Characterization Initiative (phs000235), Non-Hodgkin lymphoma and whole exome sequencing of diffuse large B-cell lymphoma (phs000450), developed by the National Cancer Institute. Some of the data used for these analyses are available from dbGAP accessions phs000235.v14.p2, phs000527.v3.p1, and phs000450.v1.p1. Information about CGCI projects can be found at https://ocg.cancer.gov/programs/cgci. We acknowledge the ICGC MALY-DE project (https://dcc.icgc.org) for providing access to their data sets. All data were used according to the data use agreements.
Authorship
Contribution: R.D.M. and K.D. performed the analysis and generated the figures and tables; and K.D., R.D.M., and P.C.B. wrote the text.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ryan D. Morin, Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada; e-mail: rdmorin@sfu.ca.
References
Author notes
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal