TO THE EDITOR:
Clinically tolerable, permissive HLA-DPB1 mismatches defined by the T-cell epitope (TCE) model improve the selection of unrelated donors in allogeneic hematopoietic cell transplantation (HCT).1 Nonpermissive mismatches across TCE groups have been shown to be associated with stronger alloreactive responses and worse clinical outcomes compared with permissive mismatches within the same TCE group.2-4 We have recently demonstrated that the biological basis of permissiveness is associated with the peptide repertoires (immunopeptidomes) presented by these molecules, which play a central role in determining the strength and T-cell receptor diversity of the alloreactive responses that they elicit.5 Less immunogenic HLA-DP molecules have similar bound peptide motifs6,7 and overlapping immunopeptidomes.5 Although the role of structural similarity and overlapping immunopeptidomes in vitro is clear,5 their relevance for alloresponses in vivo is still unknown. We hypothesized that a similarity measure reflecting the peptide-binding region of HLA-DPB1 alleles could constitute a proxy for immunopeptidome overlap and hence improve the prediction of permissive mismatches in the clinical setting.
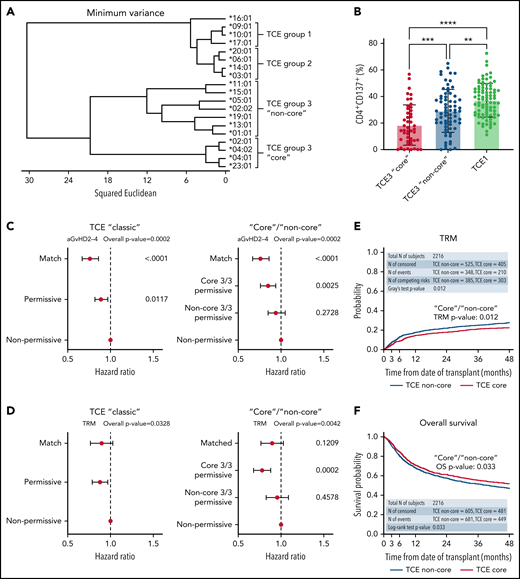
To test this hypothesis, we investigated the structural hierarchies of HLA-DPB1 alleles found in a Center for International Blood and Marrow Transplant Research (CIBMTR) cohort of 5140 10/10-matched patients who received transplants for acute myeloid leukemia, acute lymphoblastic leukemia, or myelodysplastic syndromes from 2008 to 2017 and their unrelated donors. Detailed information on the patient cohort and clinical data (supplemental Table 1, available on the Blood Web site), HLA typing and DPB1 matching, multidimensional scaling and structure analysis of HLA-DPB1 alleles, functional testing of HLA-DP alloreactive responses, mismatch stratification models, and their association with clinical outcomes can be found in the supplemental Materials. The structural relationship between HLA-DPB1 alleles was characterized by multidimensional scaling techniques based on 28 polymorphic amino acid positions encompassing all hypervariable regions in the HLA-DP molecule (supplemental Table 2). A total of 51 different HLA-DPB1 alleles, including 5 TCE group 1 (TCE1), 9 TCE2, 36 TCE3 alleles, and 1 null allele (the latter in a single patient), were identified in the clinical cohort. Clustering by amino acid sequence analysis (Figure 1A) revealed that DPB1 alleles segregate into two main branches, one including all TCE1 and TCE2 alleles and another formed by TCE3 alleles. Within TCE3, we identified two sub-branches, one formed by a subgroup of 4 frequent (cumulative allele frequencies in patients and their donors 65.0% and 66.6%, respectively) and structurally as well as functionally closely related alleles (ie, DPB1*02:01, 04:01, 04:02, 23:01). These “core” TCE3 alleles have been shown to have similar bound-peptide motifs6,7 and overlapping immunopeptidomes.5,6 Using in vitro assays, we show that TCE3 “core” alleles elicit significantly weaker (mean response 18.5%) CD4+ T-cell alloreactive responses from permissive donors compared with common “non-core” alleles (mean response 29.2%; P < .001; Figure 1B), demonstrating the functional relevance of the observed clustering. Using principal coordinates analysis, we confirmed the hypothesis that TCE3 alleles can also be distinguished by a dimorphism (DEAV/GGPM motif) formed by amino acids 84 to 87 in pocket 1,8,9 which, due to their different physicochemical properties, has been shown to have an important role in defining the peptide repertoire bound by the HLA-DP molecule10,11 (supplemental Figure 1).
Clustering analysis based on HLA-DP polymorphic positions reveals structurally and functionally divergent “core” and “non-core” HLA-DPB1 alleles predictive of clinical outcome. (A) Amino acid variation at 28 polymorphic positions (amino acids 8-215) in HLA-DPB1 coding sequences was used to cluster alleles according to their structural similarity. TCE3 “core” alleles (ie, DPB1*02:01, 04:01, 04:02, 23:01) form a distinct cluster separate from other alleles in this TCE group. For clarity, only the 19 most common alleles in the cohort, with cumulative frequencies of 98.7% and 98.8% in the patients and donors, respectively, are shown. (B) Mean in vitro alloreactive responses (% CD4+CD137+) from self-TCE3 “core” healthy donors are lowest (18.5% ± 15.2%; n = 47) against permissive TCE3 “core” alloantigens (DPB1*02:01, 04:01, 04:02) and maximal (37.2% ± 12.7%; n = 85) against nonpermissive TCE1 alleles (DPB1*09:01, 10:01, 17:01). Mean alloresponses against representative “non-core” TCE3 alleles (DPB1*01:01, 05:01, 15:01) are intermediate (29.2% ± 16.2%; n = 69) between these 2 extremes. Bars indicate mean with standard deviation. **P < .01; ***P < .001; ****P < .0001. A total of 187/201 cultures were included in Meurer et al13 and reanalyzed for this work. (C-D) Forest plots show the HR and 95% CI for (C) aGVHD II-IV and (D) TRM for the DP matching subgroups in the “classic” TCE model and the TCE3 “core” vs “non-core” stratification strategy (model II). HLA-DP nonpermissively mismatched pairs are shown as reference and overall P values of the adjusted models are presented. For statistically significant models (overall P < .01), P values for individual groups are also indicated. (E-F) Cumulative incidence (top panels) and Kaplan-Meier (bottom panels) estimates for (E) TRM and (F) overall survival in the cohort are plotted for the TCE3 “core” and “non-core” permissive subgroups. Results from statistical comparison of the curves with Gray’s test and log-rank are indicated.
Clustering analysis based on HLA-DP polymorphic positions reveals structurally and functionally divergent “core” and “non-core” HLA-DPB1 alleles predictive of clinical outcome. (A) Amino acid variation at 28 polymorphic positions (amino acids 8-215) in HLA-DPB1 coding sequences was used to cluster alleles according to their structural similarity. TCE3 “core” alleles (ie, DPB1*02:01, 04:01, 04:02, 23:01) form a distinct cluster separate from other alleles in this TCE group. For clarity, only the 19 most common alleles in the cohort, with cumulative frequencies of 98.7% and 98.8% in the patients and donors, respectively, are shown. (B) Mean in vitro alloreactive responses (% CD4+CD137+) from self-TCE3 “core” healthy donors are lowest (18.5% ± 15.2%; n = 47) against permissive TCE3 “core” alloantigens (DPB1*02:01, 04:01, 04:02) and maximal (37.2% ± 12.7%; n = 85) against nonpermissive TCE1 alleles (DPB1*09:01, 10:01, 17:01). Mean alloresponses against representative “non-core” TCE3 alleles (DPB1*01:01, 05:01, 15:01) are intermediate (29.2% ± 16.2%; n = 69) between these 2 extremes. Bars indicate mean with standard deviation. **P < .01; ***P < .001; ****P < .0001. A total of 187/201 cultures were included in Meurer et al13 and reanalyzed for this work. (C-D) Forest plots show the HR and 95% CI for (C) aGVHD II-IV and (D) TRM for the DP matching subgroups in the “classic” TCE model and the TCE3 “core” vs “non-core” stratification strategy (model II). HLA-DP nonpermissively mismatched pairs are shown as reference and overall P values of the adjusted models are presented. For statistically significant models (overall P < .01), P values for individual groups are also indicated. (E-F) Cumulative incidence (top panels) and Kaplan-Meier (bottom panels) estimates for (E) TRM and (F) overall survival in the cohort are plotted for the TCE3 “core” and “non-core” permissive subgroups. Results from statistical comparison of the curves with Gray’s test and log-rank are indicated.
Based on these observations, we postulated that HLA-DPB1 mismatches involving structurally distant alleles within TCE3 could have reduced immunopeptidome similarity and hence be less permissive than those involving structurally close alleles. To investigate this, we stratified TCE3 permissive mismatches (N = 2216) in the HCT cohort into “core” (N = 930) and “non-core” (N = 1286) or into DEAV/GGPM-matched (N = 1209) and mismatched (N = 1007) pairs and compared them with HLA-DPB1–matched (N = 785) and nonpermissively mismatched (N = 2023) pairs (supplemental Figure 2). These stratification models were tested in parallel to the “classic” TCE model considering permissive mismatches (N = 2332) as a whole. There were no major differences in clinical variables across DPB1-matched and conventional TCE matching groups or between TCE3 subgroups (supplemental Tables 1 and 3).
In line with previous results,2,12 HCT from HLA-DPB1 nonpermissive compared with matched or permissive donors according to the “classic” TCE model (model I, supplemental Figure 2) was associated with significantly higher risks of acute graft-versus-host disease (aGVHD) grades II-IV (P < .0001) (Figure 1C; Table 1). When TCE3 permissive transplants were further stratified as “core”/“non-core” (model II, supplemental Figure 2), the risks of aGVHD II-IV increased progressively from “core” TCE3 (hazard ratio [HR] 1.12 [0.98-1.28]; P = .1012) to “non-core” TCE3-permissive (HR 1.24 [1.06-1.46]; P = .0082), and nonpermissive mismatches (HR 1.32 [1.16-1.50]; P < .0001) compared with allele-matched patients (Table 1). Similarly increasing risks were observed using the stratification based on DP84-87-matched vs mismatched pairs (supplemental Table 4). When compared with nonpermissive mismatches, only “core” (HR 0.85 [0.76-0.94]; P = .0025) and DP84-87-matched (HR 0.87 [0.79-0.96]; P = .0045) but not “non-core” (HR 0.94 [0.85-1.05]; P = .2728) nor DP84-87-mismatched (HR 0.94 [0.84-1.05]; P = .3138) permissive mismatches conferred significantly lower risks of aGVHD II-IV (Figure 1C; supplemental Figure 3). Classification of TCE3-permissive pairs according to “core” and “non-core” but not according to DP84-87 matching or with the “classic” TCE model also revealed a significant association with transplant-related mortality (TRM) (Table 1; Figure 1D-E; supplemental Figure 3). Compared with the nonpermissive mismatches, the risks of TRM were significantly lower for the “core” permissive (HR 0.78 [0.68-0.88]; P = .0002) but not for the “non-core” permissive (HR 0.95 [0.83-1.09]; P = .4578). Despite a constant difference in survival between “core” and “non-core” mismatches throughout the follow-up (HR 0.88 [0.77-1.00]; P = .046) (Figure 1F), no statistically significant associations with overall survival or any of the other clinical endpoints studied were observed for any of the investigated models (supplemental Table 5).
Multivariable regression models for association between permissive and nonpermissive DPB1 mismatches and aGVHD II-IV and TRM using the classic and “core” vs “non-core” TCE models
| Endpoints . | TCE (classic) . | . | TCE3 core vs non-core . | . | ||
|---|---|---|---|---|---|---|
| DP matching . | HR (95% CI) . | (Overall) P . | DP matching . | HR (95% CI) . | (Overall) P . | |
| aGVHD II-IV | Match | 1.00 | (<.0001) | Match | 1.00 | (.0002) |
| Permissive | 1.18 (1.03-1.35) | .0197 | Core permissive | 1.12 (0.98-1.28) | .1012 | |
| Nonpermissive | 1.32 (1.16-1.50) | <.0001 | Non-core permissive | 1.24 (1.06-1.46) | .0082 | |
| Non-permissive | 1.32 (1.16-1.50) | <.0001 | ||||
| TRM | Match | 1.00 | (.0328) | Match | 1.00 | (.0042) |
| Permissive | 0.99 (0.84-1.15) | Core permissive | 0.87 (0.72-1.05) | .1439 | ||
| Nonpermissive | 1.12 (0.97-1.30) | Non-core permissive | 1.07 (0.90-1.27) | .4482 | ||
| Nonpermissive | 1.12 (0.97-1.30) | .1209 | ||||
| Endpoints . | TCE (classic) . | . | TCE3 core vs non-core . | . | ||
|---|---|---|---|---|---|---|
| DP matching . | HR (95% CI) . | (Overall) P . | DP matching . | HR (95% CI) . | (Overall) P . | |
| aGVHD II-IV | Match | 1.00 | (<.0001) | Match | 1.00 | (.0002) |
| Permissive | 1.18 (1.03-1.35) | .0197 | Core permissive | 1.12 (0.98-1.28) | .1012 | |
| Nonpermissive | 1.32 (1.16-1.50) | <.0001 | Non-core permissive | 1.24 (1.06-1.46) | .0082 | |
| Non-permissive | 1.32 (1.16-1.50) | <.0001 | ||||
| TRM | Match | 1.00 | (.0328) | Match | 1.00 | (.0042) |
| Permissive | 0.99 (0.84-1.15) | Core permissive | 0.87 (0.72-1.05) | .1439 | ||
| Nonpermissive | 1.12 (0.97-1.30) | Non-core permissive | 1.07 (0.90-1.27) | .4482 | ||
| Nonpermissive | 1.12 (0.97-1.30) | .1209 | ||||
Models were adjusted or stratified for disease type, donor age, donor/recipient CMV match, GVHD prophylaxis, HCT-CI score, conditioning regimen, year of transplant, Karnofsky score, time from diagnosis to transplant, graft type, TBI use, and disease status as required. P values for specific groups are shown only for those models in which the overall P value was significant.
Taken together, the increased risks of aGVHD and TRM detected for permissive mismatches spanning “core” and “non-core” subgroups demonstrate that a finer stratification of mismatches that reflects the immunopeptidome divergence between alleles in this group can benefit outcome prediction and potentially improve donor selection. Moreover, these results shed light on the relationship between the TCE and other models of HLA-DP permissiveness. Of note, common “non-core” DP84-87 DEAV+ alleles in TCE3 are linked to the 3′UTR high-expression rs9277534 G allele,13,14 associating a graft-versus-host mismatch between them and a “core” allele with high risk of aGVHD according to the expression model.15,16 Furthermore, “core” allele DPB1*02:01 and “non-core” allele DPB1*05:01 are the spearhead alleles of the evolutionary model for assessment of DPB1 mismatch and GVHD risk in Japan,17 with mismatches across these 2 subgroups considered detrimental. Previously described interactions between these models and the TCE model12,16-19 can be explained by the findings presented here. Importantly, per definition, the expression12,15 and DP2-DP517 models can only be applied to a fraction of the patients.20 Conversely, a refined stratification of risk based on the structural divergence among TCE3 alleles can be applied to all patient-donor pairs, producing a comprehensive, unified model able to reflect the effects of the other models.
In conclusion, in this study we have identified a “core” group of structurally and functionally related HLA-DPB1 alleles that constitute the main drivers of associations between TCE3 permissive mismatches and reduced risk of aGVHD and TRM compared with nonpermissive mismatches. These observations provide evidence for the first time that immunopeptidome similarity, central to the mechanistic basis of HLA-DPB1 TCE-permissiveness,5 also has clinical consequences for the outcome of HCT. Prospective preferential selection of “core” permissive donors within the TCE3 group to reduce transplant complications appears feasible due to the high frequency of “core” alleles21 (supplemental Figure 4). Further studies to confirm the clinical advantage of “core” TCE3 permissive mismatches in this and other clinical settings, including haploidentical transplantation,22,23 are warranted.
Acknowledgments
This work was supported by grants from the Deutsche Knochenmarkspenderdatei (DKMS-SLS-JHRG-2021-02) (E.A.-B.) and (DKMS-SLS-MHG-2018-01) (P.C.) and from the Deutsche José Carreras Leukämie Stiftung (DJCLS 20R/2019) and the Joseph Senker Stiftung (K.F.). The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute, the National Heart, Lung, and Blood Institute, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, the Health Resources and Services Administration (HHSH250201700006C), and the Office of Naval Research (N00014-20-1-2705 and N00014-20-1-2832).
Authorship
Contribution: E.A.-B., P.C., and K.F. designed the study; E.A.-B., M.H., and T.W. performed statistical analyses; E.A.-B., P.C., K.F., S.J.L., S.R.S., and Y.-T.B. analyzed and interpreted data; E.A.-B. drafted the manuscript; and all authors participated in manuscript writing and review and provided final approval of the manuscript.
Conflict-of-interest disclosure: Additional support to the CIBMTR was provided by Be the Match Foundation, the Medical College of Wisconsin, the National Marrow Donor Program, and the following commercial entities: AbbVie, Accenture, Actinium Pharmaceuticals, Inc., Adaptive Biotechnologies Corporation, Adienne SA, Allovir, Inc., Amgen, Inc., Astellas Pharma US, bluebird bio, inc., Bristol Myers Squibb Co., CareDx, CSL Behring, CytoSen Therapeutics, Inc., Daiichi Sankyo Co., Ltd., Eurofins Viracor, DBA Eurofins Transplant Diagnostics, Fate Therapeutics, Gamida-Cell, Ltd., Gilead, GlaxoSmithKline, HistoGenetics, Incyte Corporation, Iovance, Janssen Research & Development, LLC, Janssen/Johnson & Johnson, Jasper Therapeutics, Jazz Pharmaceuticals, Inc., Kadmon, Karius, Karyopharm Therapeutics, Kiadis Pharma, Kite Pharma Inc, Kite, a Gilead Company, Kyowa Kirin International plc, Kyowa Kirin, Legend Biotech, Magenta Therapeutics, Medac GmbH, Medexus, Merck & Co., Millennium, the Takeda Oncology Co., Miltenyi Biotec, Inc., MorphoSys, Novartis Pharmaceuticals Corporation, Omeros Corporation, OncoImmune, Inc., Oncopeptides, Inc., OptumHealth, Orca Biosystems, Inc., Ossium Health, Inc, Pfizer, Inc., Pharmacyclics, LLC, Priothera, Sanofi Genzyme, Seagen, Inc., Stemcyte, Takeda Pharmaceuticals, Talaris Therapeutics, Terumo Blood and Cell Technologies, TG Therapeutics, Tscan, Vertex, Vor Biopharma, and Xenikos BV.
Correspondence: Esteban Arrieta-Bolaños, Institute for Experimental Cellular Therapy, University Hospital Essen, Hufelandstr 55, 45122 Essen, Germany; e-mail: esteban.arrieta-bolanos@uk-essen.de.
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal