In this issue of Blood, Agrawal-Singh et al1 uncover a novel mechanism by which HOXA9, a transcription factor in acute myeloid leukemia (AML), represses gene expression through interacting with the scaffold attachment factor B (SAFB) protein. These findings bring into focus the transcription repression role of HOXA9 in AML.
Transcription factors play pivotal roles in hematopoietic development, and their dysregulation through aberrant expression or translocation is a recurrent feature in AML. HOXA9 is a homeodomain-containing transcription factor of the posterior HOXA gene cluster. HOXA9 plays an important role in hematopoiesis.2 Despite the mutational heterogeneity of AML, several different genetic mutations and gene fusions lead to aberrant HOXA9 activation in AML.3 In more than half of all AML cases, HOXA9 is activated through diverse mechanisms, and this activation is associated with a poor prognosis. 3,4 In these AMLs, HOXA9 inactivation impairs leukemogenesis, thus corroborating its role in AML maintenance and making it an attractive therapeutic target.5
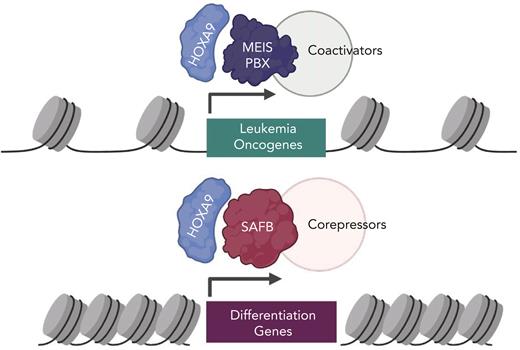
Most studies on HOXA9 activity in AML have focused on its role as a potent transcriptional activator of downstream targets that promote oncogenesis. These Hoxa9 targets include genes with prominent roles in AML such as Flt3 and Myb.3 Chromatin occupancy studies have shown that HOXA9 binds to the promoters and enhancer of its target genes, activating their expression and promoting leukemogenesis6,7 aided by recruitment of coactivators such as MLL3/4 (see figure).7,8 Interestingly, prior studies have indicated that HOXA9 may also act as a transcriptional repressor on a distinct subset of genes,6,7 but mechanisms of gene repression by HOXA9 have remained poorly studied.
A model of HOXA9 transcriptional activity in AML. (Top) HOXA9 heterodimerizes with MEIS/PBX proteins to promote activation of oncogenic signaling. Transcriptional activation may involve coactivators such as MLL3/4, proteins that modify chromatin to activate gene expression. (Bottom) A novel mechanism of HOXA9-mediated gene repression described in this study. HOXA9 interacts with the SAFB to repress differentiation-associated genes in leukemia cells. HOXA9 through SAFB recruits corepressors NuRD and HP1γ, proteins that modify chromatin to repress gene expression. Figure created with BioRender.com.
A model of HOXA9 transcriptional activity in AML. (Top) HOXA9 heterodimerizes with MEIS/PBX proteins to promote activation of oncogenic signaling. Transcriptional activation may involve coactivators such as MLL3/4, proteins that modify chromatin to activate gene expression. (Bottom) A novel mechanism of HOXA9-mediated gene repression described in this study. HOXA9 interacts with the SAFB to repress differentiation-associated genes in leukemia cells. HOXA9 through SAFB recruits corepressors NuRD and HP1γ, proteins that modify chromatin to repress gene expression. Figure created with BioRender.com.
In the article by Agrawal-Singh et al, the authors purified the HOXA9 protein interactome from an AML cell line using an endogenous HOXA9 antibody. This approach is an improvement over previous studies using HOXA9 that was ectopically overexpressed or using artificial immunoprecipitation tags. The authors identified a number of scaffold/matrix attachment region binding proteins as high-confidence HOXA9 interactors, and they focused on SAFB. The study showed that SAFB loss phenocopies HOXA9 loss in murine models of leukemia in vitro and in vivo. Interestingly, nearly one-third to two-thirds of HOXA9 binding sites were also cobound by SAFB in AML cell line and primary AML cells—demonstrating a striking degree of genomic cooccupancy. There was also a strong overlap in gene expression changes upon loss of the HOXA9 and SAFB. In contrast to need by HOXA9 to recruit coactivators for gene expression, the HOXA9/SAFB complex instead recruited the NuRD corepressor complex and the heterochromatin protein HP1γ to mediate repression of differentiation-associated genes. Targeting the catalytic components of these corepressor complexes using small molecular inhibitors phenocopied the loss of HOXA9/SAFB and showed efficacy in primary human AML patient samples. Arguably, the inhibitors used in this study, panobinostat and chaetocin, may have broader activity beyond suppression of the HOXA9/SAFB complexes; nevertheless, the results presented in this study identify a new target for reversing the repressive activity of HOXA9 in AML. Detailed characterization of HOXA9/SAFB interface will enable precise targeting of this interaction critical for HOXA9 repressive function and leukemia cell survival.
Of note, in characterizing the repressive mechanism of HOXA9 in gene regulation—and in identifying the epigenetic components that mediate this repression—Agrawal-Singh et al shed light on the mechanistic basis by which HOXA9 overexpression maintains leukemia cells in an undifferentiated state. HOXA9 overexpression has at least 2 major effects on hematopoietic cells: enhancement of self-renewal and block of differentiation. The former is relatively well understood, and the current study may help fill in the other half of the puzzle. This study also raises some thought-provoking questions. First, it is well established that HOXA9 forms heterodimers with the 3 amino-acid loop extension (TALE) domain transcription factors, including the MEIS and/or PBX proteins.3 However, it appears that HOXA9/SAFB predominantly functions independently of HOXA9/MEIS1 and has an opposite effect on transcription. Does HOXA9 form distinct, mutually exclusive complexes with the TALE proteins and with SAFB with activating and repressive functions, respectively? Second, it is not known whether similar mechanisms underly the role of HOXA9 in normal hematopoiesis. To this end, it would be interesting to investigate whether SAFB knockout in hematopoiesis has similar defects in normal hematopoietic reconstitution as Hoxa9 null hematopoietic stem cells. Finally, the exact role of SAFB in mediating HOXA9 gene repression is still enigmatic. It does not seem to affect the genomic localization of HOXA9. A previous study showed that SAFB has a role in maintaining 3D heterochromatin organization.9 This raises the interesting question whether the HOXA9/SAFB interaction serves to recruit target genes into a heterochromatic neighborhood for silencing. Future studies may help answer these questions in more detail.
Overall the Agrawal-Singh et al study identifies a novel, transcriptional repressive function for HOXA9, a transcription factor dysregulated in a large proportion of AML, opening up new avenues for therapeutic intervention in AML.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal