Key Points
Rare germline CFH variants are over-represented among patients with PNH.
Patients with PNH with rare germline CFH variants were more likely to be transfusion-dependent under eculizumab.
Abstract
Patients with paroxysmal nocturnal hemoglobinuria (PNH) are susceptible to complement-mediated intravascular hemolysis and thrombosis. Factor H (FH) is the main regulator of the complement alternative pathway, which protects cells from unwanted complement-mediated damage. Although FH is not a glycosylphosphatidylinositol-linked molecule, it may play a role in PNH. We sought to determine if rare germline variants in complement factor H (CFH) affect the PNH course, screening 84 patients with PNH treated with eculizumab for rare variants in CFH, CFI, and C3 genes. We compared the allelic frequencies with populational data and a geographically-matched control group, looking for an association between presence of the variants and treatment response (transfusion independence by 6 months). Sixteen patients presented rare variants, 9 in CFH (10.7%). Germline CFH variants were more frequent among patients with PNH than among controls (P = .02) or public data (P < .001) and were more likely to be transfusion-dependent at 6 months after eculizumab initiation (P = .015). With a median follow-up of 5.8 years, 8 of 9 patients with the CFH variant received transfusions, and 2 developed thromboses. None of the patients with the CFH variant had severe aplastic anemia from eculizumab initiation until 6 months. We demonstrated for the first time that rare CFH variants are over-represented among patients with PNH and that germline genetic background may affect the response to eculizumab.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is a rare disorder, that manifests as intravascular hemolysis, bone marrow failure and thrombosis. It is caused by somatic mutations in the PIGA gene, resulting in impaired glycosylphosphatidylinositol (GPI) biosynthesis.1 GPI anchors cell–surface proteins, including the decay-accelerating factor (CD55) and the membrane inhibitor of reactive lysis (CD59), thus modulating the alternative and terminal complement pathways. Impaired GPI synthesis leads to complement-mediated hemolysis.2 The most widely available PNH treatment, eculizumab, binds to C5, halting intravascular hemolysis and reducing thrombosis occurrence.3-5 Clinical responses vary from complete to none.6,7
Some patients may need higher doses of eculizumab to avoid residual intravascular hemolysis.8,9 C5 inhibition enhances the accumulation of C3 fragments on PNH red blood cells (RBCs), triggering extravascular hemolysis.10 Specific C5 variants are resistant to eculizumab.11 In addition, a common CR1 variant predisposes to suboptimal responses to eculizumab.12
We hypothesized that rare germline variants in regulatory genes implicated in the complement alternative pathway may influence disease presentation or response to eculizumab, notably transfusion dependence, 6 months after treatment initiation.
Study design
Data collection
We studied patients diagnosed with PNH, who were treated with eculizumab for at least 6 months and were enrolled in the French Registry of Marrow Failure Syndromes, with available samples for molecular profiling with next-generation sequencing (NGS). All participants signed informed consent. This research was carried out per the Declaration of Helsinki. The Comité de protection des personnes Ouest VI approved the research, registry, and biobank.
Gene sequencing
Genomic DNA was extracted from blood, buccal swabs, and fibroblast samples. All coding and flanking intronic sequences of CFH, CFI, MCP, and C3 genes were analyzed by NGS.13,14 Rare variants were defined as minor allelic frequency (MAF)<0.1% in the general population. Those variants in which the genetic change affected the protein function were considered pathogenic.15 Other variants were classified as being of uncertain signifcance (VUS).15,16 We confirmed the constitutional nature of the variants by sequencing DNA extracted from buccal swabs or cultured skin fibroblasts.
Statistical analyses
Qualitative and quantitative variables are presented as numbers and percentages or medians and interquartile ranges, respectively. Our cohort of patients with rare variants was compared with a control group of 80 healthy adults from France and from public data (1000 Genomes).
Transfusion dependence was the primary end point, defined as >2 RBC transfusion episodes during 6 months. The secondary outcomes were failure-free survival (FFS), defined as the first RBC transfusion after 28 days from eculizumab initiation, an increase in eculizumab dose, thrombosis, or death and event-free survival (EFS), defined as FFS also including hematopoietic cell transplantation or evolution to a WHO (2016)–defined myeloid malignancy. Symptoms of anemia or a hemoglobin level below 8 g/dL, triggered blood transfusions.
EFS and FFS were analyzed using the Kaplan-Meier estimator. The impact of variables on outcomes was tested using Fisher or Wilcoxon and log-rank tests. We present EFS and FFS with 95% confidence intervals (CI). We used the R software for statistical analyses.
Results and discussion
We screened 84 patients for genetic variants and evaluated 83 patients for primary and secondary objectives (supplemental Figure 1; available on the Blood website).
To the best of our knowledge, it is for the first time that we report the frequency of rare variants in CFH (9/84, 10.7%), CFI (3/84, 3.6%), and C3 (4/84, 4.8%) genes in patients with PNH (Table 1). We confirmed the germline nature of CFH variants in 10 patients using DNA from buccal swabs (n = 2) or fibroblasts (n = 8).
Rare germline CFH variants among patients with PNH
| UPN . | Gene . | Variant . | Transcript . | Status . | MAF (%) . | CADD score . | In vitro functional characterization23 . | Summary of the effect . | Classification in this report . |
|---|---|---|---|---|---|---|---|---|---|
| PNH016 | CFH | c.472G>A | p.Val158Ile | Hz | 0.018 | 0.001 | yes | Any identified functional defect24 | VUS |
| PNH028 | CFH | c.2077G>A | p.Asp693Asn | Hz | 0.016 | 20.8 | no | Low FH plasma level | Pathogenic |
| PNH030 | CFI | c.672T>A | p.Asp224Glu | Hz | novel | 0.038 | yes | Any identified functional defect25 | VUS |
| PNH032 | C3 | c.664T>A | p.Phe222Ile | Hz | 0.005 | 27.3 | no | VUS | |
| PNH032 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS | |
| PNH032 | CFI | c.1246A>C | p.Ile416Leu∗ | Hz | 0.02 | 18.64 | yes | Significantly reduced expression compared with WT26 | Pathogenic |
| PNH033 | CFH | c.3004G>C | p.Gly1002Arg | Hz | 0.078 | 17.45 | no | Low FH plasma level with Alternative pathway C3 consumption | Pathogenic |
| PNH037 | CFH | c.2651C>A | p.Ser884Tyr | Hz | 0.032 | 17.24 | yes | Any identified functional defect24 | VUS |
| PNH038 | CFI | c.1534G>A | p.Gly512Ser | Hz | 0.0017 | 33 | yes | Any identified functional defect25 | VUS |
| PNH046 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS | |
| PNH060 | C3 | c.749G>T | p.Gly250Val | Hz | novel | 19.09 | no | VUS | |
| PNH073 | CFH | c.1366G>A | p.Glu456Lys | Hz | 0.0008 | 2.658 | no | Normal C3 level in plasma | VUS |
| PNH078 | C3 | c.1119+1G>A | IVS10+1 | Hz | novel | 35 | no | Splice site variant | Pathogenic |
| PNH087 | CFH | c.2295A>T | p.Leu765Phe | Hz | 0.0036 | 0.035 | no | Normal C3 level in plasma | VUS |
| PNH097 | C3 | c.3083T>G | p.Leu1028Arg | Hz | novel | 26.7 | no | VUS | |
| PNH097 | CFH | c.472G>A | p.Val158Ile | Hz | 0.018 | 0.001 | yes | Any identified functional defect25 | VUS |
| PNH102 | CFH | c.2867C>T | p.Thr956Met | Hz | 0.1 | 14.99 | yes | Any identified functional defect25 | VUS |
| PNH103 | CFH | c.388G>A | p.Asp130Asn | Hz | 0.014 | 22.6 | yes | Normal C3 and FH levels in plasma, minor defects in complement regulation23 | Pathogenic |
| PNH108 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS |
| UPN . | Gene . | Variant . | Transcript . | Status . | MAF (%) . | CADD score . | In vitro functional characterization23 . | Summary of the effect . | Classification in this report . |
|---|---|---|---|---|---|---|---|---|---|
| PNH016 | CFH | c.472G>A | p.Val158Ile | Hz | 0.018 | 0.001 | yes | Any identified functional defect24 | VUS |
| PNH028 | CFH | c.2077G>A | p.Asp693Asn | Hz | 0.016 | 20.8 | no | Low FH plasma level | Pathogenic |
| PNH030 | CFI | c.672T>A | p.Asp224Glu | Hz | novel | 0.038 | yes | Any identified functional defect25 | VUS |
| PNH032 | C3 | c.664T>A | p.Phe222Ile | Hz | 0.005 | 27.3 | no | VUS | |
| PNH032 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS | |
| PNH032 | CFI | c.1246A>C | p.Ile416Leu∗ | Hz | 0.02 | 18.64 | yes | Significantly reduced expression compared with WT26 | Pathogenic |
| PNH033 | CFH | c.3004G>C | p.Gly1002Arg | Hz | 0.078 | 17.45 | no | Low FH plasma level with Alternative pathway C3 consumption | Pathogenic |
| PNH037 | CFH | c.2651C>A | p.Ser884Tyr | Hz | 0.032 | 17.24 | yes | Any identified functional defect24 | VUS |
| PNH038 | CFI | c.1534G>A | p.Gly512Ser | Hz | 0.0017 | 33 | yes | Any identified functional defect25 | VUS |
| PNH046 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS | |
| PNH060 | C3 | c.749G>T | p.Gly250Val | Hz | novel | 19.09 | no | VUS | |
| PNH073 | CFH | c.1366G>A | p.Glu456Lys | Hz | 0.0008 | 2.658 | no | Normal C3 level in plasma | VUS |
| PNH078 | C3 | c.1119+1G>A | IVS10+1 | Hz | novel | 35 | no | Splice site variant | Pathogenic |
| PNH087 | CFH | c.2295A>T | p.Leu765Phe | Hz | 0.0036 | 0.035 | no | Normal C3 level in plasma | VUS |
| PNH097 | C3 | c.3083T>G | p.Leu1028Arg | Hz | novel | 26.7 | no | VUS | |
| PNH097 | CFH | c.472G>A | p.Val158Ile | Hz | 0.018 | 0.001 | yes | Any identified functional defect25 | VUS |
| PNH102 | CFH | c.2867C>T | p.Thr956Met | Hz | 0.1 | 14.99 | yes | Any identified functional defect25 | VUS |
| PNH103 | CFH | c.388G>A | p.Asp130Asn | Hz | 0.014 | 22.6 | yes | Normal C3 and FH levels in plasma, minor defects in complement regulation23 | Pathogenic |
| PNH108 | C3 | c.4100T>C | p.Ile1367Thr | Hz | 0.081 | 17.41 | no | VUS |
Allelic frequency has been taken from the Genome Aggregation Database (gnomAD). PNH032 carried a pathogenic CFI variant (p.Ile416Leu) and 2 C3 variants of undetermined significance (p.Ile1367Thr and p.Phe222Ile ); PNH097 carried 2 VUS variants in C3 (p.Leu1028Arg) and CFH (p.Val158Ile).
CADD, combined annotation dependent depletion; Hz, heterozygous; VUS, variants of undetermined significance.
The allelic frequency for p.Ile416Leu in the African population is 1.2%.
We found that CFH variants were significantly over-represented among patients with PNH when compared with healthy controls (P = .02) or to public data (P<.001, supplemental Table 1). In contrast to CFH, the frequency of CFI or C3 variants was not increased, highlighting FH’s role in RBC protection.
FH, a central alternative pathway regulator, prevents the formation of the C3 convertase and regulates C3 convertase decay and the degradation of C3b on cells’ surfaces, acting as a cofactor to factor I.17 In PNH cells, only CR1 and FH protect RBCs against complement. FH protection is crucial for PNH cells’ survival.18 Enhanced surface levels of FH on the RBCs of CD55 or CD59–deficient mice and patients with PNH protected the RBCs against complement-mediated hemolysis.19
Based on available data, 3 out of 8 rare CFH variants were pathogenic, causing alternative pathway deregulation. These 3 unstudied variants may affect FH-cell binding. In the absence of functional studies, the relevance of identified variants in the pathophysiology of PNH remains unclear. However, the over-representation of rare variants in disease-specific populations indicates the relevance of the gene to disease pathogenesis.15,20,21
The duration of PNH diagnoses ranged from 1978 to 2019; the first and last eculizumab initiations took place in 2005 and 2019, respectively. The transfusion recommendations were stable during these 14 years (supplemental Table 1). Two patients started eculizumab, having relapsed for PNH after hematopoietic stem cell transplantation for aplastic anemia with autologous reconstitution. Twenty-seven patients received 1200 mg (instead of 900 mg) of eculizumab owing to suboptimal responses, of which 3 had a rare CFH variant.
Compared with wild-type, patients bearing a CFH variant were older at PNH diagnosis; other baseline characteristics were not different. During follow-up, the results of the diagnoses of 7 patients evolved to a myeloid malignancy. None of these patients had a rare variant in the studied genes.
The CFH variants or transfusion requirements before eculizumab initiation were associated with transfusion dependence at 6 months after eculizumab initiation in univariate analysis (75.7% vs 33.3% for CFH variant and wild-type, respectively, P = .015, supplemental Table 3); no other baseline characteristics affected transfusion dependence. All 3 patients with pathogenic CFH variants and 1 with pathogenic CFI were transfusion-dependent for 6 months (supplemental Table 5).
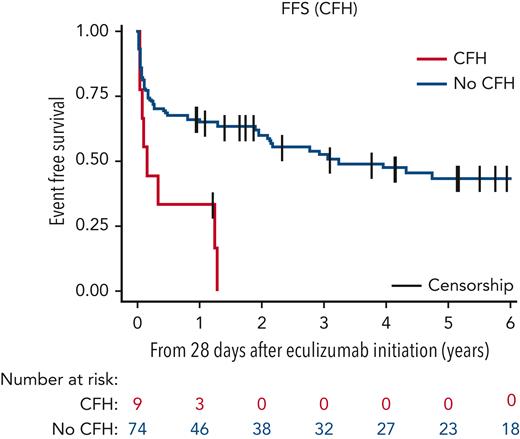
Median follow-up was 5.8 years (95% CI, 5.2-7.3). Six-year FFS and EFS were identical because transfusions preceded other events as 39.3% (95% CI, 28.1-50.3) and 0% for wild-type and CFH-variant groups, respectively (Figure 1; supplemental Figure 3; supplemental Table 4). All but 1 patient with CFH variant received transfusions after eculizumab loading dose (PNH037 had a VUS and remained transfusion-independent [supplemental Table 5]; eculizumab started after a thrombotic event). Two patients with CFH variant had a first thrombosis under eculizumab therapy. None of the patients with CFH variant fulfilled the criteria for severe aplastic anemia after eculizumab initiation. Overall, patients younger in age at PNH diagnosis, without a CFH variant, and transfusion independent before eculizumab treatment had a better 6-year EFS (supplemental Table 4).
FFS of patients with PNH bearing a rare CFH variant (red) vs without a CFH variant (blue). The 6-year FFS for the no-CFH group was 39.3 months (95% CI, 28.1-50.3). Events included, first red blood cell transfusion after the eculizumab loading dose, thrombosis, eculizumab dose increase to 1200 mg, or occurrence of death.
FFS of patients with PNH bearing a rare CFH variant (red) vs without a CFH variant (blue). The 6-year FFS for the no-CFH group was 39.3 months (95% CI, 28.1-50.3). Events included, first red blood cell transfusion after the eculizumab loading dose, thrombosis, eculizumab dose increase to 1200 mg, or occurrence of death.
We did not find any association among common variants in CR1 (associated with lower protein expression) or in CFH p.His402Tyr (implicated in other complement-mediated diseases)22 with disease presentation or eculizumab response (supplemental Table 3; supplemental Table 6; supplemental Table 7; supplemental Figure 4).
Our data support that FH protection is necessary for the survival of PNH cells, even during C5 cleavage inhibition. Decreased FH regulation on RBCs under C5 inhibition would further amplify C3 fragment–deposition on PNH cell surfaces, enhancing extravascular hemolysis and contributing to poor eculizumab responses. We did not observe striking differences in hemoglobin or lactate dehydrogenase between patients with or without a rare CFH variant (supplemental Figure 2) from day 28 to 6 months. These observations do not favor intravascular hemolysis as the cause for the suboptimal response to anti-C5 treatment, but suggest a more indolent evolution.
The limitations of this study were the retrospective design, the possibility of referral bias (national reference center), and the lack of subgroup or multivariate analyses owing to the disease rarity. Our results require an independent validation.
Patients bearing these CFH variants may respond differently to proximal complement inhibitors. Screening for CFH variants may help clinicians understand the suboptimal response to terminal inhibition and possibly adapt treatment with new proximal complement inhibitors.
Acknowledgments
This study was partly supported by research funding from the Association HPN France – Aplasie Médullaire and Fondation Maladies Rares and the American Society of Hematology Global Research Award to P.H.P.
Authorship
Contribution: P.H.P. helped in designing the project, participated in patient recruitment, collected, analyzed, interpreted clinical and molecular data, and wrote the manuscript; J.-E.G. performed statistical analysis on clinical data; F.S.d.F., R.B., and Y.B. participated in patient recruitment and collected samples; P.-E.D. participated in patient recruitment and collected clinical data; P.V.M, A.D., and S.R. performed molecular analyses; A.-C.L. collected clinical data; L.L. and J.S. prepared and biobanked samples and gave helpful intellectual insights; G.S. participated in patient recruitment, gave helpful intellectual insights, and edited the manuscript; V.F.-B. performed molecular analyses, interpreted molecular data, helped elaborate and sponsor the project, and edited the manuscript; R.P.d.L. helped in designing and sponsoring the project, participated in patient recruitment, analyzed, interpreted clinical and molecular data, and edited the manuscript; and all authors read and approved the final version of the manuscript and are accountable for all aspects of the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pedro Henrique Prata, Hematology and Transplantation, Saint-Louis Hospital, 1 Avenue Claude Vellefaux, 75010 Paris, France; e-mail: pedrohenrique.delimaprata@aphp.fr; and Véronique Frémeaux-Bacchi, Laboratoire d’Immunologie Biologique, Hôpital Européen Georges Pompidou, 20 Rue Leblanc, 75015 Paris, France; e-mail: veronique.fremeaux-bacchi@aphp.fr.
References
Author notes
∗V.F.-B. and R.P.d.L. contributed equally to this study.
Data (anonymized) are available upon request from the corresponding authors, Pedro Henrique Prata (pedrohenrique.delimaprata@aphp.fr) and Véronique Frémeaux-Bacchi (veronique.fremeaux-bacchi@aphp.fr).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal