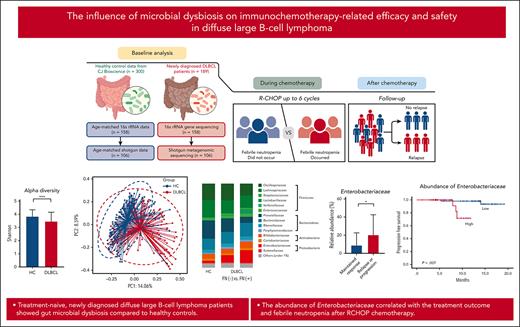
Key Points
Treatment-naive, newly diagnosed patients with DLBCL exhibited gut microbial dysbiosis.
The abundance of Enterobacteriaceae correlated with treatment outcome and febrile neutropenia after RCHOP chemotherapy.
Abstract
The gut microbiome influences cancer development and the efficacy and safety of chemotherapy but little is known about its effects on lymphoma. We obtained stool samples from treatment-naive, newly diagnosed patients with diffuse large B-cell lymphoma (DLBCL) (n = 189). We first performed 16S ribosomal RNA gene sequencing (n = 158) and then conducted whole-genome shotgun sequencing on additional samples (n = 106). We compared the microbiome data from these patients with data from healthy controls and assessed whether microbiome characteristics were associated with treatment outcomes. The alpha diversity was significantly lower in patients with DLBCL than in healthy controls (P < .001), and the microbial composition differed significantly between the groups (P < .001). The abundance of the Enterobacteriaceae family belonging to the Proteobacteria phylum was markedly higher in patients than in healthy controls. Functional analysis of the microbiome revealed an association with opportunistic pathogenesis through type 1 pili, biofilm formation, and antibiotics resistance. Enterobacteriaceae members were significantly enriched in patients who experienced febrile neutropenia and in those who experienced relapse or progression (P < .001). Interestingly, greater abundance of Enterobacteriaceae correlated with shorter progression-free survival (P = .007). The cytokine profiles of patients whose microbiome was enriched with Enterobacteriaceae were significantly associated with interleukin 6 (P = .035) and interferon gamma (P = .045) levels. In summary, patients with DLBCL exhibited gut microbial dysbiosis. The abundance of Enterobacteriaceae correlated with treatment outcomes and febrile neutropenia. Further study is required to elucidate the origin and role of gut dysbiosis in DLBCL.
Introduction
The gut microbiota has an important influence on the development and homeostasis of the host immune system.1-3 Compositional and functional changes in the gut microbiota, called gut dysbiosis, may lead to epithelial barrier dysfunction and alterations in innate and adaptive immunity.4 This may lead to gut dysbiosis-related chronic inflammation, which may play a role in malignancy, and gut dysbiosis may impair the efficacy and safety of anticancer treatments.5-8 Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin lymphoma (NHL) and requires immediate treatment because of its aggressive clinical features such as multiple progressive lymphadenopathies and extranodal involvement.9,10 Although the role of the gut microbiota has not been elucidated fully in patients with DLBCL, a previous study using 16S ribosomal RNA (rRNA) gene and whole-genome shotgun (WGS) sequencing showed that most patients with DLBCL exhibit microbial dysbiosis that involves members of the Proteobacteria.11 However, whether the gut microbiota influences treatment outcomes of patients with DLBCL is unclear.
Once diagnosed with DLBCL, patients should be treated with cytotoxic chemotherapy combined with B-cell–depleting anti-CD20–targeting antibodies such as the combination therapy rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone (RCHOP).12 Although RCHOP is widely used as the standard treatment for patients with DLBCL, ∼40% of patients with DLBCL cannot be cured because of their repeated relapses or refractoriness to treatments.3 Febrile neutropenia is one of the most common treatment-related adverse events after RCHOP chemotherapy, especially in older patients, and the incidence of febrile neutropenia after RCHOP is ∼20%.13-15 Because febrile neutropenia can lead to treatment delay, dose reduction, and discontinuation of RCHOP, prophylaxis with pegylated granulocyte colony–stimulating factor (G-CSF) is recommended for patients receiving RCHOP.16
A recent study using 16S rRNA gene sequencing of DLBCL or follicular lymphomas found that microbial dysbiosis is associated with aggressive histology and adverse clinical outcomes.17 However, little has been published on the effects of the gut microbiota on febrile neutropenia after RCHOP chemotherapy in patients with DLBCL. Considering that RCHOP confers a risk of febrile neutropenia, and that bacteria in the gastrointestinal tract can act as pathogens, analysis of the gut microbiota in patients with DLBCL at the time of DLBCL diagnosis may provide important information for the management of patients who will receive RCHOP chemotherapy. In this study, we collected stool samples from consecutive treatment-naive patients diagnosed with DLBCL and compared the gut microbiota of these patients with that of healthy controls. We used 16S rRNA and WGS sequencing metagenomics to evaluate the composition of the gut microbiota, and analyzed the association with the occurrence of febrile neutropenia and treatment outcomes after RCHOP chemotherapy.
Materials and methods
Cohort study
We collected stool samples prospectively from treatment-naive patients newly diagnosed with DLBCL who were registered in our prospective cohort study, between 2019 and 2021 (NCT 03117036). The inclusion criteria were as follows: (1) patients newly diagnosed (treatment naive) with DLBCL who were scheduled to receive immunochemotherapy, and (2) patients aged ≥20 years. The exclusion criteria were as follows: (1) the presence of inflammatory bowel disease, (2) a history of acute inflammatory or infectious disease within 1 month before study participation, (3) use of antibiotics, steroids, or immunosuppressants within 1 month before study participation, (4) prior surgery involving the gastrointestinal tract, and (5) use of artificial nutrition. All patients provided written informed consent in accordance with the institutional review board regulations before participating in this study.
The following pretreatment clinical and laboratory information was recorded: sex, age, complete blood count, serum lactate dehydrogenase (LDH) and β-2 microglobulin levels, Eastern Cooperative Oncology Group (ECOG) performance status, body mass index, bone marrow involvement, Ann Arbor stage and International Prognostic Index (IPI), National Comprehensive Cancer Network IPI, and central nervous system IPI.18,19 Each patient’s pathology results were reviewed by a lymphoma pathologist (J.H.), and the cell of origin was identified at the time of diagnosis.
Patients were treated with RCHOP chemotherapy for up to 6 cycles, and pegylated G-CSF was administered prophylactically in every cycle. The occurrence of febrile neutropenia was documented during RCHOP chemotherapy and defined as a case requiring hospitalization or a visit to the emergency room because of a febrile episode related to neutropenia, which was defined as an absolute neutrophil count of <1000 cells per mm3 with a single temperature measurement of >38.3°C or a sustained temperature of ≥38°C for <1 hour. The response evaluation was conducted in clinical practice after the third and sixth cycles and was based on the Lugano response criteria.20 Patients responding to RCHOP chemotherapy were monitored regularly for surveillance of relapse or progression. Information about each patient’s disease status and survival was updated prospectively in this cohort, and the cutoff date for this study was 30 June 2022. This study was approved by the institutional review board of Samsung Medical Center and performed under all relevant guidelines and regulations, including those of the Declaration of Helsinki.
Stool sample collection and DNA extraction
Based on to the inclusion and exclusion criteria, a total of 189 patients were enrolled in this study. Most patients collected their own stool samples using a sampling kit at home using the following method: fresh stools were obtained aseptically before the onset of RCHOP chemotherapy using a stool sampling kit containing buffer for storing the stool samples at room temperature (CJ Bioscience Inc, Seoul, Korea). Samples were shipped to CJ Bioscience, and DNA was extracted for 16S rRNA analysis (supplemental Methods, available on the Blood website).
Analysis of the gut microbiota
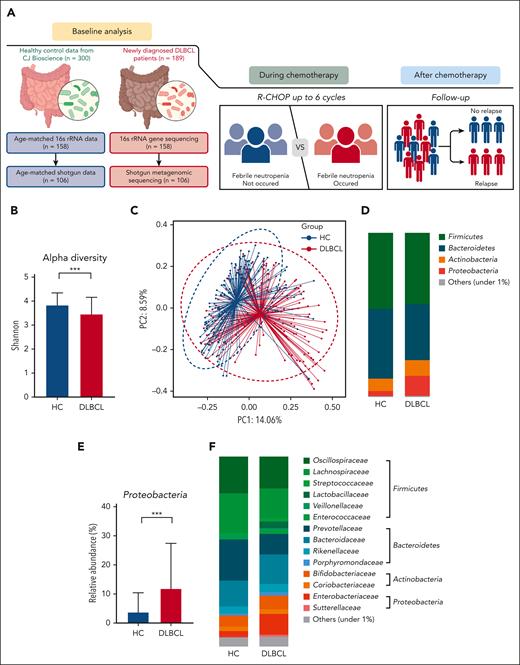
Of the 189 participants from whom stool samples were collected, we performed 16S rRNA gene sequencing on stool samples from 158 patients whose sample quality was appropriate for the analysis. Their gut microbiota, based on the results of 16S rRNA gene sequencing, was compared with that of age- and sex-matched healthy controls (n = 158). For functional and species-level taxonomic analysis, additional WGS sequencing was performed on samples from 106 of those 158 patients, from whom stool samples were available for additional shotgun sequencing (Figure 1A).
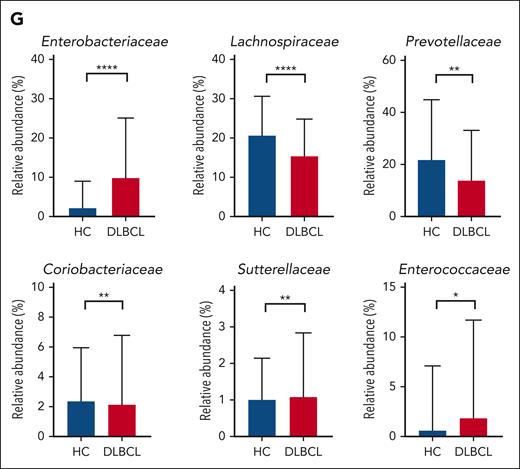
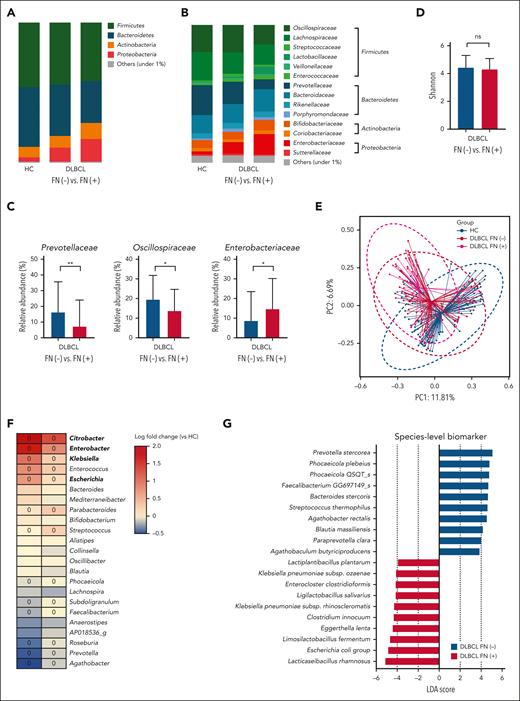
Comparison of gut microbiota between patients with DLBCL and healthy controls. (A) Description of the study. (B-C) Comparisons of alpha diversity using the Shannon index, and beta diversity using the Bray-Curtis index between patients with DLBCL and healthy controls. (D-E) Comparisons of the relative abundance at the phylum level between patients with DLBCL and healthy controls. (F-G) Comparisons of the relative abundance at the family level between patients with DLBCL and healthy controls. Asterisks indicate significant differences identified using the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. HC, healthy controls; ns, not significant.
Comparison of gut microbiota between patients with DLBCL and healthy controls. (A) Description of the study. (B-C) Comparisons of alpha diversity using the Shannon index, and beta diversity using the Bray-Curtis index between patients with DLBCL and healthy controls. (D-E) Comparisons of the relative abundance at the phylum level between patients with DLBCL and healthy controls. (F-G) Comparisons of the relative abundance at the family level between patients with DLBCL and healthy controls. Asterisks indicate significant differences identified using the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. HC, healthy controls; ns, not significant.
The first objective of our analysis was to identify the presence of gut dysbiosis in patients with DLBCL compared with healthy controls. The second objective was to determine whether the microbiome data obtained from 16S rRNA and shotgun sequencing at diagnosis were associated with the occurrence of febrile neutropenia during RCHOP chemotherapy. The third objective was to determine whether the characteristics of the gut microbiota were associated with treatment outcomes by comparing these in patients who relapsed or progressed after RCHOP chemotherapy with those whose response was maintained (Figure 1A).
16S rRNA gene sequencing and taxonomic profiling
The V3-4 hypervariable region of the 16S rRNA gene was amplified with primers 341F and 805R using the direct polymerase chain reaction method. Libraries were prepared using a NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA). The prepared DNA libraries were sequenced by CJ Bioscience Inc using the Illumina MiSeq platform (Illumina, San Diego, CA) with 2 × 300 base pair (bp) kits (supplemental Methods).
WGS metagenomic sequencing
The WGS sequencing metagenomic libraries were prepared using DNA stored in a previous step and NEBNext Ultra II DNA Library Prep Kit and NEBNext Multiplex Oligos for Illumina (New England Biolabs), per the manufacturer’s protocols. Fragment size and DNA concentration in the final library were checked using a Bioanalyzer system (Agilent Technologies, Santa Clara, CA) and then sequenced using an Illumina NovaSeq 6000 platform (2 × 150 bp read length) at Macrogen (Seoul, Korea).
Taxonomic profiling of shotgun metagenomics data
A Kraken2 database21 containing bacterial and archaeal species represented in the EzBioCloud database was generated,22 and 92 core genes were extracted using the UBCG pipeline.23 The total core gene length for each species was stored for further downstream analysis. A Kraken2-compatible taxonomic structure was constructed using EzBioCloud’s taxonomic system, and the core gene sequences were converted into FASTA files using a numerical identifier matching the taxonomic structure file. Finally, the database was compiled with the Kraken2 “build” command, with k = 35 and default parameters. The potential presence of bacterial and archaeal species for each raw metagenomic sample read was initially surveyed using the prebuilt Kraken2 core-gene database.24 After acquiring a list of candidate species, a custom Bowtie2 database was built using only the core genes from the species found during the first step to reduce the search space and obtain accurate coverage and depth metrics.25 The raw sample was then mapped against the Bowtie2 database using the compassionate option and a quality threshold of phred33. SAMtools was used to convert and sort the output BAM file.26 Coverage of the mapped reads against the BAM file was obtained using BEDtools.27 To avoid false positives, reads that mapped to a given species were quantified only if the total coverage of their core genes was at least 25%, per an in-house script. Finally, species abundance was calculated using the total number of reads counted, and normalized species abundance was calculated using the total length of core genes per species.
Functional profiling based on shotgun metagenomics data
Functional annotations were obtained by matching each read against the KEGG database28 using DIAMOND.29 An initial database file was built from the KEGG FASTA file containing all the ortholog amino acid sequences using DIAMOND’s “makedb” command with the default parameters. DIAMOND was then executed using the BLASTx parameter, which converts each metagenomic read into multiple amino acid sequences by generating all 6 open reading frames and then matching these against the prebuilt KEGG database. If a read had multiple KEGG hits, the top hit was used. After quantifying all KEGG orthologs, MinPath was used to predict the presence of KEGG functional pathways.30
Machine learning for discriminating DLBCL
Using the WGS sequencing data for the gut microbiome, we developed a random forest classifier for discriminating DLBCL from healthy control samples using custom Python scripts and the Scikit-learn package.31 We trained random forest classifiers with bacterial composition (gut = species and genus level) and function (gene and pathway level). The model was trained 20 times using a fivefold cross-validation method, and the average area under the receiver operating characteristic curve was calculated.
Multiplex cytokine analysis
In an additional study to explore the patients’ pretreatment inflammatory state, we measured pretreatment cytokine levels in serum samples obtained at the time of stool sampling. The levels of cytokines interferon gamma (IFN-γ), interleukin-1 receptor antagonist (IL-1RA), IL-1β, IL-4, IL-6, IL-10, IL-12, IL-13, IL-17α, IL-18, IL-23, tumor necrosis factor α (TNF-α), and TNF-β were analyzed using a Procarta cytokine profiling kit (Panomics, Fremont, CA) and the Bio-Plex cytokine assay system (Bio-Rad Laboratories, Hercules, CA) per the manufacturers’ instructions. Cytokines levels were also analyzed with regards to the gut microbiome composition and clinical information.
Statistical analysis
The χ2 test, Student t test, and Mann-Whitney U test were applied, where appropriate, to compare the baseline demographics using IBM SPSS Statistics (version 25.0; IBM Corp, Armonk, NY). The species richness was assessed using Chao1, and diversity indices were calculated using the Shannon matrix. Beta diversity was calculated using the Bray-Curtis metric and visualized with principal coordinate analysis. The significance of beta diversity was assessed using permutational multivariate analysis of variance, and microbial taxonomy was assigned by the Quantitative Insights into Microbial Ecology 2.32
To examine the taxonomic and functional differences between patients with DLBCL and healthy controls, we performed linear discriminant analysis effect size (LEfSe) analysis.33 The variation in each taxonomic profile and differences in function were analyzed using the Mann-Whitney U test. Using XLSTAT software (Addinsoft, Paris, France), we performed canonical correspondence analysis. Correlations between the abundance of gut bacteria and blood cytokine levels were calculated using Spearman rank correlation analysis. Network maps were generated using Quantitative Insights Into Microbial Ecology 2 SCNIC34 and visualized in Cytoscape version 3.8.2.35 Progression-free survival (PFS) was defined as the time from the date of diagnosis to the date of disease progression, relapse, death due to any cause, or the last follow-up. Survival was estimated based on Kaplan-Meier curves and compared using log-rank tests. Two-sided P values <.05 were considered significant. Cox hazard regression analysis was conducted for multivariate analysis of the association between PFS and microbiome data.
Results
Patient characteristics and outcomes
Among the 189 patients, more men (n = 119, 63%) than women (n = 70, 37%) were enrolled, and 91 patients were aged >60 years (48%, Table 1). Most patients (n = 144, 76%) had ECOG performance status scores of 0 and therefore did not require hospitalization. Elevated serum LDH levels were found in 90 patients (48%), and only 12 patients had bone marrow involvement (6%). Stage 3/4 accounted for 42% of patients (n = 80). Based on the IPI score, 95 patients were classified as having a low IPI risk (score of 0 or 1) and 19 patients as being at high risk (Table 1).
Patient characteristics and outcomes
| . | Total (N = 189) . | 16S rRNA (n = 158) . | Shotgun (n = 106) . |
|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |
| Baseline characteristics | |||
| Sex | |||
| Male | 119 (63) | 101 (64) | 63 (59) |
| Female | 70 (37) | 57 (36) | 43 (41) |
| Age, y | |||
| ≤60 | 98 (52) | 83 (53) | 61 (58) |
| >60 | 91 (48) | 75 (47) | 45 (42) |
| BMI, kg/m2 | |||
| <25 | 125 (66) | 104 (66) | 70 (66) |
| ≥25 | 64 (34) | 54 (34) | 36 (34) |
| ECOG PS | |||
| 0 | 144 (76) | 121 (76) | 78 (74) |
| 1 | 44 (23) | 37 (23) | 28 (25) |
| 2 | 1 (1) | 1 (1) | 1 (1) |
| B symptoms | |||
| Presence | 7 (4) | 5 (3) | 4 (4) |
| Absence | 182 (96) | 153 (97) | 102 (96) |
| Serum LDH | |||
| Elevated | 90 (48) | 74 (47) | 47 (44) |
| Normal | 99 (52) | 84 (53) | 59 (56) |
| β2-microglobulin | |||
| Elevated | 30 (16) | 23 (15) | 16 (15) |
| Normal | 137 (72) | 122 (77) | 80 (76) |
| Not examined | 22 (12) | 13 (8) | 10 (9) |
| CRP | |||
| Elevated | 73 (39) | 61 (39) | 38 (36) |
| Normal | 96 (51) | 81 (51) | 54 (51) |
| Not examined | 20 (10) | 16 (10) | 14 (13) |
| Cell of origin | |||
| ABC | 102 (54) | 83 (53) | 57 (54) |
| GCB | 61 (32) | 54 (34) | 36 (34) |
| Not determined | 26 (14) | 21 (13) | 13 (12) |
| Stage | |||
| I/II | 109 (58) | 94 (60) | 57 (54) |
| III/IV | 80 (42) | 64 (40) | 49 (46) |
| Bone marrow | |||
| Involved | 12 (6) | 11 (7) | 9 (9) |
| Not involved | 155 (82) | 132 (84) | 84 (79) |
| Not examined | 22 (12) | 15 (9) | 13 (12) |
| IPI | |||
| Low | 95 (50) | 83 (53) | 55 (52) |
| Low-intermediate | 40 (21) | 34 (21) | 23 (22) |
| High-intermediate | 35 (19) | 27 (17) | 17 (16) |
| High | 19 (10) | 14 (9) | 11 (10) |
| Treatment outcome after RCHOP | |||
| Complete response | 171 (90) | 146 (92) | 101 (95) |
| Partial response | 9 (5) | 8 (5) | 5 (5) |
| Progression | 4 (2) | 4 (3) | |
| Not evaluated | 5 (3) | ||
| Febrile neutropenia | 39 (21) | 30 (19) | 23 (22) |
| Disease relapse during follow-up | 20 (11) | 17 (11) | 11 (10) |
| . | Total (N = 189) . | 16S rRNA (n = 158) . | Shotgun (n = 106) . |
|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |
| Baseline characteristics | |||
| Sex | |||
| Male | 119 (63) | 101 (64) | 63 (59) |
| Female | 70 (37) | 57 (36) | 43 (41) |
| Age, y | |||
| ≤60 | 98 (52) | 83 (53) | 61 (58) |
| >60 | 91 (48) | 75 (47) | 45 (42) |
| BMI, kg/m2 | |||
| <25 | 125 (66) | 104 (66) | 70 (66) |
| ≥25 | 64 (34) | 54 (34) | 36 (34) |
| ECOG PS | |||
| 0 | 144 (76) | 121 (76) | 78 (74) |
| 1 | 44 (23) | 37 (23) | 28 (25) |
| 2 | 1 (1) | 1 (1) | 1 (1) |
| B symptoms | |||
| Presence | 7 (4) | 5 (3) | 4 (4) |
| Absence | 182 (96) | 153 (97) | 102 (96) |
| Serum LDH | |||
| Elevated | 90 (48) | 74 (47) | 47 (44) |
| Normal | 99 (52) | 84 (53) | 59 (56) |
| β2-microglobulin | |||
| Elevated | 30 (16) | 23 (15) | 16 (15) |
| Normal | 137 (72) | 122 (77) | 80 (76) |
| Not examined | 22 (12) | 13 (8) | 10 (9) |
| CRP | |||
| Elevated | 73 (39) | 61 (39) | 38 (36) |
| Normal | 96 (51) | 81 (51) | 54 (51) |
| Not examined | 20 (10) | 16 (10) | 14 (13) |
| Cell of origin | |||
| ABC | 102 (54) | 83 (53) | 57 (54) |
| GCB | 61 (32) | 54 (34) | 36 (34) |
| Not determined | 26 (14) | 21 (13) | 13 (12) |
| Stage | |||
| I/II | 109 (58) | 94 (60) | 57 (54) |
| III/IV | 80 (42) | 64 (40) | 49 (46) |
| Bone marrow | |||
| Involved | 12 (6) | 11 (7) | 9 (9) |
| Not involved | 155 (82) | 132 (84) | 84 (79) |
| Not examined | 22 (12) | 15 (9) | 13 (12) |
| IPI | |||
| Low | 95 (50) | 83 (53) | 55 (52) |
| Low-intermediate | 40 (21) | 34 (21) | 23 (22) |
| High-intermediate | 35 (19) | 27 (17) | 17 (16) |
| High | 19 (10) | 14 (9) | 11 (10) |
| Treatment outcome after RCHOP | |||
| Complete response | 171 (90) | 146 (92) | 101 (95) |
| Partial response | 9 (5) | 8 (5) | 5 (5) |
| Progression | 4 (2) | 4 (3) | |
| Not evaluated | 5 (3) | ||
| Febrile neutropenia | 39 (21) | 30 (19) | 23 (22) |
| Disease relapse during follow-up | 20 (11) | 17 (11) | 11 (10) |
ABC, activated B cell; BMI, body mass index; CRP, C-reactive protein; GCB, germinal-center B cell; PS, performance status.
The overall response rate to RCHOP chemotherapy was 95% (180 of 189) and included 171 patients with a complete response. In total, 4 patients were refractory to RCHOP, and no response evaluation was performed in 5 patients. Febrile neutropenia occurred in 39 patients (21%) during RCHOP chemotherapy. At the median follow-up of 16.4 months (95% confidence interval, 15.1-17.7 months), disease relapse occurred in 20 patients (Table 1). The gut microbiota was first examined using 16S rRNA gene sequencing in 158 of the 189 patients, and the WGS sequencing was performed in the 106 of these 158 patients for whom samples for shotgun sequencing were available. Baseline characteristics and treatment outcomes after RCHOP chemotherapy for these patients were similar to those for the total study population (Table 1).
Comparison of gut microbiota between patients with DLBCL and healthy controls
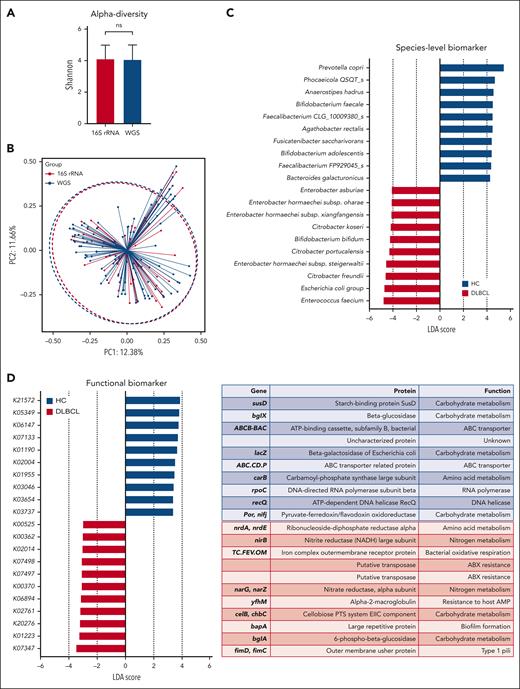
The 16S rRNA gene sequencing showed that patients with DLBCL had lower alpha diversity than did the healthy controls, and that beta diversity also differed significantly between patients with DLBCL and healthy controls (P < .001; Figures 1B-C). In the taxonomic comparison of gut microbial composition at the phylum level, Proteobacteria was significantly more abundant in patients with DLBCL than in healthy controls (Figures 1D-E). At the family level, patients with DLBCL had a greater abundance of Enterobacteriaceae and Sutterellaceae, whereas healthy controls showed a greater abundance of Lachnospiraceae, Prevotellaceae, and Coriobacteriaceae (Figures 1F-G). WGS sequencing performed as functional- and species-level taxonomic analysis in 106 patients showed no significant differences in the gut microbiota between analysis using 16S rRNA and WGS sequencing (Figures 2A-B). Using the WGS sequencing data, LEfSe analysis identified the abundance of facultatively anaerobic species, especially those of the Enterobacteriaceae family, including Escherichia coli, Citrobacter freundii, and Enterobacter faecium at the species level (Figure 2C). The community functional analysis showed an association of these anaerobic species with opportunistic pathogenesis, such as type 1 pili, biofilm formation, host antimicrobial peptide resistance, and antibiotics resistance genes (Figure 2D). By contrast, Prevotella, Phocaeicola, Anaerostipes, Bifidobacterium, Faecalibacterium, and Agathobacter, which belong to the short-chain fatty acids (SCFA)-producing commensal bacterial group, were abundant in the healthy controls (Figure 2C).
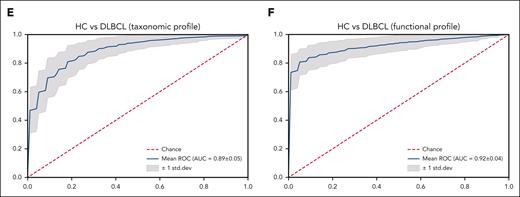
Functional- and species-level taxonomic analysis. (A-B) Comparisons of alpha diversity using the Shannon index, and beta diversity using the Bray-Curtis index between the data for 16S rRNA (n = 158) and WGS sequencing (n = 106). (C) LEfSe analysis of species-level taxonomic biomarkers for DLBCL. (D) Findings of the community functional analysis in patients with DLBCL. The top 10 features with the highest linear discriminant analysis scores in each group are shown. (E-F) Machine learning analysis to examine the applicability of microbiome data to the species-level taxonomic profile and the gene-level functional profile in patients with DLBCL and in healthy controls. AMP, antimicrobial peptide; HC, healthy controls.
Functional- and species-level taxonomic analysis. (A-B) Comparisons of alpha diversity using the Shannon index, and beta diversity using the Bray-Curtis index between the data for 16S rRNA (n = 158) and WGS sequencing (n = 106). (C) LEfSe analysis of species-level taxonomic biomarkers for DLBCL. (D) Findings of the community functional analysis in patients with DLBCL. The top 10 features with the highest linear discriminant analysis scores in each group are shown. (E-F) Machine learning analysis to examine the applicability of microbiome data to the species-level taxonomic profile and the gene-level functional profile in patients with DLBCL and in healthy controls. AMP, antimicrobial peptide; HC, healthy controls.
Machine learning analysis was conducted to validate the feasibility of this microbiome database for distinguishing people with DLBCL from the healthy population. The training model using the random forest method through the species-level taxonomic and gene-level functional profiles of patients with DLBCL showed high prediction accuracy in terms of the taxonomic profile (Figure 2E) and the functional profile (Figure 2F).
Microbiome dysbiosis and febrile neutropenia
Of the 106 patients whose samples were analyzed using WGS sequencing, 23 experienced febrile neutropenia during RCHOP chemotherapy (Table 1); the dosage was reduced in 16 of these 23 patients. Patients experiencing febrile neutropenia had higher levels of Proteobacteria at the phylum level than those who did not but this difference was not statistically significant (Figure 3A). However, the comparison at the family level showed an abundance of Enterobacteriaceae in patients with febrile neutropenia, whereas those without febrile neutropenia showed an abundance of Prevotellaceae and Oscillospiraceae (Figures 3B-C). These results suggest that the abundance of Enterobacteriaceae differs between patients with DLBCL who do and do not develop febrile neutropenia.
Microbiome dysbiosis and febrile neutropenia. (A-B) Comparisons of the relative abundance at the phylum and family levels between patients who did and did not experience febrile neutropenia and healthy controls. (C) Comparisons of the relative abundance at the family level between patients who did and did not experience febrile neutropenia. (D-E) Alpha and beta diversity analysis of the characteristics of patients with and without febrile neutropenia and healthy controls. (F) Heat map shows genera with significant differences between healthy controls and patients with DLBCL. The range of colors indicates log-fold changes in genera abundance from that observed in the healthy controls, with a color range from red, which indicates greater abundance in patients with DLBCL, to blue, which indicates less abundance in patients with DLBCL. Boxes marked with a circle show a significant difference between patients who did and did not experience febrile neutropenia. (G) At the species level, the LEfSe analysis shows the abundance of Escherichia coli and Klebsiella pneumoniae in patients with febrile neutropenia. Asterisks indicate significant differences identified using the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. FN, febrile neutropenia; HC, healthy controls; ns, not significant.
Microbiome dysbiosis and febrile neutropenia. (A-B) Comparisons of the relative abundance at the phylum and family levels between patients who did and did not experience febrile neutropenia and healthy controls. (C) Comparisons of the relative abundance at the family level between patients who did and did not experience febrile neutropenia. (D-E) Alpha and beta diversity analysis of the characteristics of patients with and without febrile neutropenia and healthy controls. (F) Heat map shows genera with significant differences between healthy controls and patients with DLBCL. The range of colors indicates log-fold changes in genera abundance from that observed in the healthy controls, with a color range from red, which indicates greater abundance in patients with DLBCL, to blue, which indicates less abundance in patients with DLBCL. Boxes marked with a circle show a significant difference between patients who did and did not experience febrile neutropenia. (G) At the species level, the LEfSe analysis shows the abundance of Escherichia coli and Klebsiella pneumoniae in patients with febrile neutropenia. Asterisks indicate significant differences identified using the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. FN, febrile neutropenia; HC, healthy controls; ns, not significant.
Although the alpha diversity did not differ significantly with regards to the presence of febrile neutropenia (P = .211; Figure 3D), the beta diversity differed between patients with and without febrile neutropenia as well as healthy controls for both comparisons (P < .001; Figure 3E). This finding shows that the patients with DLBCL who developed febrile neutropenia during RCHOP chemotherapy had a microbiome profile with a greater abundance of Enterobacteriaceae compared with the healthy controls as well as patients with DLBCL without febrile neutropenia. Linear regression analysis showed that the main driving force for the difference in beta diversity was the abundance of Enterobacteriaceae (Figure 3F). At the species level, the LEfSe analysis identified the abundance of E coli and Klebsiella pneumoniae in patients with febrile neutropenia (Figure 3G). Functional analysis of these species also demonstrated a relationship with type 1 pili, fimbriae, and antibiotic resistance in patients with febrile neutropenia (supplemental Table 1).
Microbiome dysbiosis and treatment outcomes
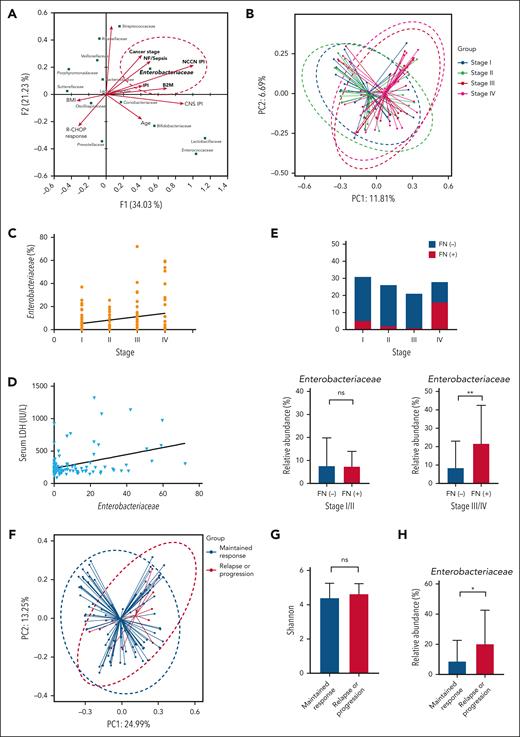
Canonical correspondence analysis was conducted to explore the relationships between bacterial species and the clinical and laboratory features reflecting the tumor burden. Of all bacterial families, only Enterobacteriaceae was significantly associated with the features of high tumor burden, such as stage 3 or 4 disease (Figure 4A). Comprehensive prognostic indexes, such as the IPI and National Comprehensive Cancer Network–IPI, also correlated significantly with Enterobacteriaceae abundance, although age and central nervous system–IPI did not. Stage correlated significantly with the abundance of Enterobacteriaceae in the linear regression (Figure 4A). Comparison of beta diversity based on the stage also showed a significant difference in gut microbiota between stages 1 or 2 and 3 or 4 (P < .001; Figure 4B).
Microbiome dysbiosis and treatment outcomes. (A) Canonical correspondence analysis to identify specific characteristics in patients with DLBCL. (B) Bray-Curtis analysis of beta diversity shows differences between patients with DLBCL with limited stages and those with advanced stages of disease. (C-D) Stage and serum LDH level were associated with the abundance of Enterobacteriaceae. (E) Patients with stage IV experienced febrile neutropenia and patients with advanced-stage disease and febrile neutropenia had a greater abundance of Enterobacteriaceae. (F-H) Comparisons of beta diversity using the Bray-Curtis index, alpha diversity using the Shannon index, and relative abundance of Enterobacteriaceae based on disease relapse or progression. Asterisks indicate significant differences identified with the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. ns, not significant.
Microbiome dysbiosis and treatment outcomes. (A) Canonical correspondence analysis to identify specific characteristics in patients with DLBCL. (B) Bray-Curtis analysis of beta diversity shows differences between patients with DLBCL with limited stages and those with advanced stages of disease. (C-D) Stage and serum LDH level were associated with the abundance of Enterobacteriaceae. (E) Patients with stage IV experienced febrile neutropenia and patients with advanced-stage disease and febrile neutropenia had a greater abundance of Enterobacteriaceae. (F-H) Comparisons of beta diversity using the Bray-Curtis index, alpha diversity using the Shannon index, and relative abundance of Enterobacteriaceae based on disease relapse or progression. Asterisks indicate significant differences identified with the 2-tailed Mann-Whitney U test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. ns, not significant.
The abundance of Enterobacteriaceae showed an association with the patients’ stage (P = .035; Figure 4C) and serum LDH level (P = .002; Figure 4D). Patients with stage 3 or 4 disease experienced febrile neutropenia more frequently than those with stage 1 or 2 (P < .001; Figure 4E). In the subgroup analysis based on stage, the abundance of Enterobacteriaceae was significantly enriched in patients with stage 3 or 4 (P = .004; Figure 4E). Despite the association between microbiome dysbiosis and tumor burden, alpha diversity did not differ significantly with regards to the occurrence of relapse or progression (P = .467; Figure 4F). However, beta diversity (P = .043; Figure 4G) and relative abundance of Enterobacteriaceae (P = .038; Figure 4H) differed significantly between patients who relapsed or progressed and those whose response was maintained.
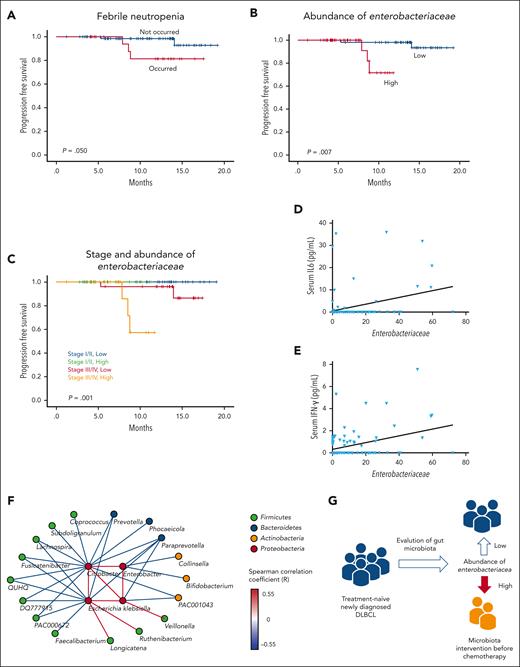
In the survival analysis, patients who experienced febrile neutropenia showed significantly worse PFS than those who did not have febrile neutropenia during RCHOP chemotherapy (P = .050; Figure 5A). After dichotomization of the 106 patients whose samples were analyzed by WGS sequencing into low (0-50th percentile) and high (51-100th percentile) abundance of Enterobacteriaceae, patients with high abundance of Enterobacteriaceae had significantly worse PFS than did those with low abundance (P = .007; Figure 5B). Patients with stage 3 or 4 disease and high abundance of Enterobacteriaceae had the worst PFS (P = .001; Figure 5C). However, the PFS did not differ between patients with low abundance of Enterobacteriaceae with stage 3 or 4 disease and those with stage 1 or 2 disease. The baseline characteristics did not differ significantly between patients with low and high abundance of Enterobacteriaceae (supplemental Table 2). The multivariate analysis of survival showed that the abundance of Enterobacteriaceae was independently associated with poor PFS (Table 2).
Survival analysis of DLBCL patients. (A-B) Comparison of PFS based on the presence or absence of febrile neutropenia and the abundance of Enterobacteriaceae. (C) Comparison of PFS based on stage and abundance of Enterobacteriaceae. (D-E) Associations of the abundance of Enterobacteriaceae with the serum levels of IL-6 and IFN-γ. (F) Network analysis of genera belonging to Enterobacteriaceae and significantly correlated genera. The color of the nodes indicates the phylum to which they belong, and the color of the edge indicates the Spearman correlation coefficient between nodes. (G) A suggested adjuvant prophylactic treatment plan based on the microbiome for preventing adverse events in patients with DLBCL.
Survival analysis of DLBCL patients. (A-B) Comparison of PFS based on the presence or absence of febrile neutropenia and the abundance of Enterobacteriaceae. (C) Comparison of PFS based on stage and abundance of Enterobacteriaceae. (D-E) Associations of the abundance of Enterobacteriaceae with the serum levels of IL-6 and IFN-γ. (F) Network analysis of genera belonging to Enterobacteriaceae and significantly correlated genera. The color of the nodes indicates the phylum to which they belong, and the color of the edge indicates the Spearman correlation coefficient between nodes. (G) A suggested adjuvant prophylactic treatment plan based on the microbiome for preventing adverse events in patients with DLBCL.
Univariate and multivariate analyses of PFS
| . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Male | 1.643 | 0.473-5.701 | .434 | 1.817 | 0.478-6.905 | .381 |
| Age >60 y | 1.374 | 0.398-4.746 | .616 | 0.707 | 0.169-2.959 | .635 |
| IPI risk | 1.724 | 1.090-2.729 | .020 | 1.751 | 0.318-9.649 | .520 |
| Febrile neutropenia | 3.616 | 1.045-12.505 | .042 | 2.070 | 0.461-9.299 | .343 |
| Abundance of Enterobacteriaceae | 13.730 | 1.704-110.624 | .014 | 11.905 | 1.424-99.542 | .022 |
| . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Male | 1.643 | 0.473-5.701 | .434 | 1.817 | 0.478-6.905 | .381 |
| Age >60 y | 1.374 | 0.398-4.746 | .616 | 0.707 | 0.169-2.959 | .635 |
| IPI risk | 1.724 | 1.090-2.729 | .020 | 1.751 | 0.318-9.649 | .520 |
| Febrile neutropenia | 3.616 | 1.045-12.505 | .042 | 2.070 | 0.461-9.299 | .343 |
| Abundance of Enterobacteriaceae | 13.730 | 1.704-110.624 | .014 | 11.905 | 1.424-99.542 | .022 |
CI, confidence interval; HR, hazard ratio.
Because pretreatment inflammatory cytokines may be associated with outcomes, we evaluated the associations between gut microbiota and pretreatment cytokine levels. In the samples obtained at the time of stool sampling, serum IL-6 (P = .035) and IFN-γ (P = .045) levels were significantly associated with Enterobacteriaceae abundance (Figures 5D and E).
Discussion
The gut microbiota is a complex community comprising trillions of microbes in the gastrointestinal tract and is known to influence aspects of host immunity such as regulatory T-cell generation and natural killer function.36,37 Imbalances in the gut microbiota may lead to abnormal immune status and the development of cancers.38 Associations between microbiota composition and anticancer treatment outcomes have been reported and include effects on the response to cytotoxic chemotherapy as well as immune checkpoint inhibitors.39,40 A recent multicenter observational study that assessed fecal microbiota composition and diversity found that patients with lymphoma, myeloma, or amyloidosis with lower fecal diversity around the time of neutrophil engraftment had higher rates of disease progression after autologous stem cell transplantation.41 In another study, previous exposure to antibiotics that alter the gut microbiome was significantly associated with clinical outcomes in patients with NHL receiving CD19-targeted chimeric antigen receptor T-cell therapy, which suggests a role of the fecal microbiome in the responses to chimeric antigen receptor T-cell immunotherapy.42
In this study, we collected baseline stool samples prospectively from newly diagnosed, treatment-naive patients with DLBCL and evaluated the relationships between the gut microbiota and treatment outcome and the presence of febrile neutropenia after RCHOP chemotherapy. After taxonomic profiling using 16S rRNA gene sequencing (n = 158), we conducted WGS sequencing (n = 106) of the baseline stool samples obtained before the first cycle of RCHOP chemotherapy to explore species-level taxonomic and gene-level functional biomarkers. Comparison of the gut microbiota between patients with DLBCL and healthy controls showed that patients had significantly lower alpha diversity and different beta diversity. We also found differences in bacterial compositions between these groups, including a high abundance of Enterobacteriaceae and Sutterellaceae in patients with DLBCL and an abundance of Lachnospiraceae, Prevotellaceae, and Coriobacteriaceae in healthy controls. In the high-dimensional class comparisons using LEfSe, patients with DLBCL had greater relative abundance of facultatively anaerobic species, including E coli, C freundii, and E faecium at the species level. The differences in the microbiome found in baseline stool samples suggest that patients with DLBCL could be distinguished from the healthy population using the taxonomic and function profile in the machine learning analysis.
Based on these findings, we hypothesize that gut dysbiosis may be involved in the development of DLBCL. However, various other factors, such as lifestyle, may influence the gut microbiota. Although we excluded patients with inflammatory or infectious disease as well as those who used antibiotics, steroids, or immunosuppressants within 1 month before participation in this study, a broad range of external factors may have influenced the detected alterations in the microbiota observed in these patients. The presence of DLBCL itself may lead indirectly to gut dysbiosis because of changes in lifestyle, such as dietary changes because of a loss of appetite after the development of DLBCL. Therefore, we cannot conclude that there is a causal relationship between gut dysbiosis and DLBCL based only on our current findings. Considering the limitations of our study, further studies are needed to identify the origin of the gut dysbiosis in patients with DLBCL.
Comparison of the fecal microbiome composition in patients with DLBCL based on the occurrence of febrile neutropenia failed to show a significant difference in alpha diversity. However, different beta diversity was found in patients with DLBCL who experienced febrile neutropenia compared with those who did not and healthy controls. LEfSe analysis showed that the main difference in beta diversity related to the abundance of Enterobacteriaceae, and E coli and K pneumoniae at the species level. These organisms are common pathogens in patients with febrile neutropenia and are frequently related to the occurrence of shock and early death during chemotherapy.43,44 The functional analysis showed a relationship between these organisms and the formation of type 1 pili, metabolism of carbohydrates and nitrogen, and resistance to antibiotics. Type 1 pilus–mediated bacterial invasion and host-pathogen interactions have been reported in various clinical situations.45,46
Gut microbial dysbiosis may also lead to abnormal immune status through the overproduction of oxidative stress and inflammatory cytokines.47-49 To determine the pretreatment inflammatory status, we measured baseline cytokine concentrations in serum samples obtained along with stool samples before the onset of RCHOP chemotherapy. Although the statistical differences were modest, Enterobacteriaceae abundance was significantly associated with IL-6 (P = .035) and IFN-γ (P = .045) levels in patients enriched with these organisms (Figures 5D-E). Thus, the detected alterations in the gut microbiota may be associated with pretreatment inflammatory status considering that serum LDH level may reflect the extent of inflammation and predict poor prognosis in patients with DLBCL.50 However, increased serum levels of inflammatory cytokines could be caused by various factors other than the gut microbiome, and additional functional studies are warranted to elucidate the mechanism underlying the interaction between the gut microbiota and inflammation in patients with DLBCL. Such studies should include metabolomics analysis of both stool and serum samples because metabolites derived from the microbiome may play an essential role in the host.
Despite the high cure rate in patients treated with RCHOP, the risk of infection-related mortality remains challenging, especially for older patients.51,52 Because the occurrence of febrile neutropenia may be associated with comorbidity, performance status, and chemotherapy-induced hematological toxicity, physicians must sometimes reduce the dosage of chemotherapy, even in the first cycle. This change may interfere with the ability to deliver sufficient amounts of medication, which may lead to progression or relapse, and may explain the poor outcome of older patients with DLBCL. Patients who experience febrile neutropenia may be more likely to require a reduced dosage of chemotherapy and to experience delayed treatment, which can adversely affect the treatment outcome of DLBCL.
In this study, we found frequent occurrence of disease progression and poor PFS in patients enriched in Enterobacteriaceae. Therefore, we propose a supportive adjuvant strategy to restore a healthy intestinal microbial community by treating the consortia of commensal bacteria that compete with Enterobacteriaceae for the ecological niche and to restore colonization resistance against Enterobacteriaceae. The network analysis in our study identified Faecalibacterium, Prevotella, Paraprevotella, and Bifidobacterium as commensal bacteria whose presence correlated negatively with the 4 genera that accounted for most of the Enterobacteriaceae in the intestine of these patients, namely Escherichia, Citrobacter, Klebsiella, and Enterobacter (Figure 5F). These well-known beneficial strains produce SCFA in the intestine, which reduces gut permeability and promotes aerobic respiration in colonocytes to remove intestinal oxygen. Through its high metabolic efficiency in aerobic respiration, Enterobacteriaceae abundance may threaten the survival of other organisms. Therefore, increasing the abundance of other strains identified here may help in the deoxygenation of the intestinal environment and reduction of the abundance of Enterobacteriaceae (Figure 5G).
In conclusion, to our knowledge, this is the first comprehensive microbiome study to use WGS sequencing in the analysis of the microbiome of treatment-naive patients with DLBCL. Our comparison of the microbiome between patients with DLBCL and healthy controls found that patients with DLBCL developed intestinal dysbiosis that involved a greater abundance of Enterobacteriaceae. We also found that patients with advanced-stage disease who experienced treatment-related febrile neutropenia were more likely to have abundant Enterobacteriaceae, and that patients with enriched Enterobacteriaceae had poor outcomes. Considering these results, we suggest increasing the community of commensal bacteria to restore a healthy intestinal microbial community to reduce the abundance of Enterobacteriaceae. However, more extensive clinical studies are needed to validate these findings. The possible role of the gut microbiota in the occurrence of febrile neutropenia and its influence on treatment outcomes should be clarified to improve the outcomes of patients with DLBCL receiving RCHOP chemotherapy.
Acknowledgments
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (HR20C0025); a National Research Foundation of Korea grant funded by the Korean government (2022R1F1A1064058); and a grant from the International Congress of Korean Society of Hematology (ICKSH) Research Project through the Korean Society of Hematology, Republic of Korea (grant number: ICKSH-2022-05).
Authorship
Contribution: S.J.K., S.E.Y., and W.S.K. designed the research; S.E.Y., W.K., S.C., Y.P., M.C., H.K., J.H.L., D.-W.H., H.S., K.J.R., J.-Y.L., and J.-W.B. performed the research; W.S.K. and S.J.K. collected patient clinical data; S.J.K., S.E.Y., and W.K. analyzed the data and wrote the manuscript; and all authors edited the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seok Jin Kim, Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, 81 Irwon-ro Gangnam-gu, Seoul, 06351, Korea; e-mail: kstwoh@skku.edu.
References
Author notes
∗S.E.Y. and W.K. contributed equally to this study as joint first authors.
The datasets generated during the current study are available in the NCBI repository (https://www.ncbi.nlm.nih.gov) and Sequence Read Archive database under the accession numbers PRJNA928211. Data are available upon request from the authors (starload0326@gmail.com).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal