Key Points
Arginine metabolism regulates erythroid lineage differentiation of human progenitor cells via eIF5A-induced protein synthesis.
RP haploinsufficiency negatively affects eIF5A hypusination in erythroid progenitors.
Abstract
Metabolic programs contribute to hematopoietic stem and progenitor cell (HSPC) fate, but it is not known whether the metabolic regulation of protein synthesis controls HSPC differentiation. Here, we show that SLC7A1/cationic amino acid transporter 1–dependent arginine uptake and its catabolism to the polyamine spermidine control human erythroid specification of HSPCs via the activation of the eukaryotic translation initiation factor 5A (eIF5A). eIF5A activity is dependent on its hypusination, a posttranslational modification resulting from the conjugation of the aminobutyl moiety of spermidine to lysine. Notably, attenuation of hypusine synthesis in erythroid progenitors, by the inhibition of deoxyhypusine synthase, abrogates erythropoiesis but not myeloid cell differentiation. Proteomic profiling reveals mitochondrial translation to be a critical target of hypusinated eIF5A, and accordingly, progenitors with decreased hypusine activity exhibit diminished oxidative phosphorylation. This affected pathway is critical for eIF5A-regulated erythropoiesis, as interventions augmenting mitochondrial function partially rescue human erythropoiesis under conditions of attenuated hypusination. Levels of mitochondrial ribosomal proteins (RPs) were especially sensitive to the loss of hypusine, and we find that the ineffective erythropoiesis linked to haploinsufficiency of RPS14 in chromosome 5q deletions in myelodysplastic syndrome is associated with a diminished pool of hypusinated eIF5A. Moreover, patients with RPL11-haploinsufficient Diamond-Blackfan anemia as well as CD34+ progenitors with downregulated RPL11 exhibit a markedly decreased hypusination in erythroid progenitors, concomitant with a loss of mitochondrial metabolism. Thus, eIF5A-dependent protein synthesis regulates human erythropoiesis, and our data reveal a novel role for RPs in controlling eIF5A hypusination in HSPCs, synchronizing mitochondrial metabolism with erythroid differentiation.
Introduction
Hematopoietic stem cells (HSCs) possess 2 fundamental characteristics: the capacity for self-renewal and the sustained production of all blood cell lineages. The differentiation of HSCs to the erythroid lineage is distinctive in that progressive mitoses result in the production of daughter cells that differ, morphologically and functionally, from their parent cell, with terminal erythroid differentiation generating enucleated reticulocytes.
The maturation of erythroid precursors is regulated by transcription factors, cell-cell contacts, oxygen sensors, and cytokines, most notably, erythropoietin (EPO).1-6 Notably though, erythropoiesis is also a formidable process at a metabolic level. The production of 200 × 107 red cells per day is highly dependent on iron, glucose, fatty acid, and amino acid metabolism. Amino acids contribute to multiple aspects of erythroblast metabolism, ranging from precursors of nucleic acids, mTOR sensing, production of tricarboxylic acid cycle intermediates, and maintenance of intracellular redox, among others.7-15 Arginine, a semiessential amino acid,16 affects erythroid progenitors17,18 but its function in erythroid differentiation is unknown.
Arginine catabolism results in the generation of a diverse range of products, including nitric oxide, urea, creatine, proline, glutamate, agmatine, and polyamines, including spermidine. Of note, the spermidine polyamine is required for hypusination of eukaryotic translation initiation factor 5A (eIF5A). After hypusination, eIF5A promotes translation elongation as well as translation termination.19-21 Here, we show that the spermidine-dependent hypusination of eIF5A is required for erythroid lineage commitment as well as for terminal erythroid differentiation. The critical nature of protein synthesis in erythropoiesis is highlighted by the finding that mutations in ribosomal protein (RP) genes underlie the hypoplastic anemia associated with Diamond-Blackfan anemia (DBA) and myelodysplastic syndromes (MDSs) with chromosome 5q deletions [del(5q)].22,23 In light of our results showing the importance of eIF5A-stimulated translation in erythroid differentiation, we evaluated the impact of RP haploinsufficiency on hypusination. Strikingly, our data identify a novel link between RPs and hypusination in the regulation of erythropoiesis: haploinsufficiency of RPs significantly decreased hypusinated eIF5A (eIF5AH) in erythroid progenitors. Thus, our study reveals a mechanism via which RPs regulate hypusination, thereby governing erythroid differentiation.
Methods
CD34+ selection
All studies involving human samples were conducted in accordance with the declaration of Helsinki. CD34+ cells were isolated after informed consent and approval by the respective institutional review boards. Samples from patients with and without MDS were obtained from the HemoDiag prospective cohort of patients with hemopathy (#NCT02134574, Centre Hospitalier Universitaire de Montpellier). CD34+ cells were differentiated as detailed in supplemental Methods, which is available on the Blood website.
Lentiviral transduction
Flow cytometry
Metabolic assays
Mitochondrial biomass and activity were monitored by flow cytometry; mitochondrial and glycolysis stress tests were performed on an XFe-96 Analyzer (Seahorse Bioscience, Agilent); and glutamine and glucose uptake were evaluated using radioactively labeled substrates, as detailed in the supplemental Methods.
Protein synthesis analyses
Protein synthesis rate was monitored by the Click-iT Plus OPP Protein Synthesis Assay, as described by the manufacturer (Molecular Probes).
Western blots
After transfer of protein gels to polyvinylidene fluoride, membranes were incubated with 5% nonfat dry milk, incubated with the indicated antibodies, and followed by peroxidase anti-rabbit or anti-mouse immunoglobulin, as described in the supplemental Methods.
Proteomics profiling
EPO-stimulated CD34+ cells were treated with N1-guanyl-1,7-diaminoheptane (GC7) (5 μM) for 48 hours. Cells from 3 donors were washed, and dry pellets were conserved at −80 °C until extraction (supplemental Methods).
Results
EPO-stimulated human progenitors exhibit significantly attenuated erythroid but not myeloid differentiation in the absence of SLC7A1-mediated arginine transport
The availability of intracellular arginine is regulated in large part by the cell-surface expression of the SLC7 cationic amino acid transporter (CAT) family.26,27 Erythroid differentiation of human CD34+ hematopoietic stem and progenitor cells (HSPCs), induced by recombinant EPO (supplemental Figure 1A-C), results in a significant upregulation of SLC7A1 (P < .05) (Figure 1A), as well as arginine uptake (P < .05) (Figure 1B; supplemental Figure 1D). Interestingly, this increase in arginine transport does not represent a global augmentation in amino acid uptake; rEPO stimulation did not augment transport of glutamine (supplemental Figure 1D), an amino acid critical for erythroid development.10,11 Cell-surface SLC7A1 levels then decreased during terminal erythroid differentiation (P < .001) (Figure 1C), associated with a fivefold reduction in arginine uptake from days 4 to 10 of differentiation (P < .01) (Figure 1D). During differentiation, arginine was transported into these cells by SLC7A1/CAT1 and not y+LAT/4F2hc;28 N-ethylmaleimide, a pharmacological agent that blocks CAT1 but not y+LAT,29 inhibited arginine uptake to levels detected in red blood cells (P < .0001) (supplemental Figure 1E).
HSC specification to the erythroid lineage is dependent on the expression and function of the SLC7A1 arginine transporter. (A) Cell-surface expression of SLC7A1 was evaluatedafter rEPO-induced erythroid differentiation of CD34+ progenitors (day 4) and representative histograms in the absence or presence of rEPO are presented (left). Relative MFIs of SCL7A1 in rEPO-induced progenitors are compared with levels detected in the absence of EPO (right, n = 5 independent experiments). (B) Arginine uptake in the absence or presence of rEPO (day 4) was monitored using L-[2,3,4-3H)] arginine monohydrochloride (2 μCi) for 10 minutes at room temperature. Uptake in the presence of EPO was arbitrarily set at 1 (means of triplicates in 3 independent experiments). (C) Surface SLC7A1 levels were monitored at days 4, 7, and 10 of rEPO-mediated differentiation and representative histograms are shown (left). Mean fluorescent intensities (MFIs) of SLC7A1 staining relative to day 4 were quantified (right, n = 9). (D) Arginine uptake was evaluated at days 4, 7, and 10 of erythroid differentiation; arginine uptake at day 4 was arbitrarily set at 1 (means of triplicates in 4 independent experiments). (E) CD34+ progenitors were transduced 3 days with GFP-tagged shCTRL and shSLC7A1 lentiviral vectors and representative histograms of cell-surface SLC7A1 expression on GFP+ cells (left) as well as quantification of SLC7A1 expression relative to control-transduced cells is shown (right, n = 8 independent experiments). (F) Arginine uptake was monitored in fluorescence-activated cell sorter–sorted progenitors transduced with shCTRL and shSLC7A1 vectors at day 4 and uptake levels relative to the shCTRL condition are presented (n = 3). (G) The evolution of shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of GFP expression at days 0, 3, and 7 of rEPO-induced differentiation and representative histograms are shown (left). Quantification of the percentages of GFP+ cells relative to day 0 is presented (right, n = 6). (H) Differentiation of rEPO-induced shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of CD34 and CD36 expression on IL3R−GlyA− cells; CFU-E are defined by an IL3R−GlyA−CD34−CD36+ phenotype (left, day 3). Quantification of CFU-E relative to shCTRL-transduced progenitors is presented (right, n = 9). (I) The differentiation of progenitors to an erythroid (GlyA) vs myeloid (CD11b) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative plots (left) and quantifications (right) are presented. Surface expression of GlyA and CD11b was monitored on shCTRL- and shSLC7A1-transduced progenitors at day 3 of differentiation (left). Quantification of erythroid (n = 14) and myeloid (n = 12) differentiation is shown in independent experiments (right). (J) CD34+CD38− progenitors transduced with shCTRL and shCAT1 vectors were evaluated for CFU potential using the StemMACS HSC-CFU Assay Kit and the numbers of burst-forming unit erythroid (BFU-E) and CFU–granulocyte-macrophage (GM) colonies generated at day 14 are presented for 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. IgG, immunoglobulin G; ns, not significant.
HSC specification to the erythroid lineage is dependent on the expression and function of the SLC7A1 arginine transporter. (A) Cell-surface expression of SLC7A1 was evaluatedafter rEPO-induced erythroid differentiation of CD34+ progenitors (day 4) and representative histograms in the absence or presence of rEPO are presented (left). Relative MFIs of SCL7A1 in rEPO-induced progenitors are compared with levels detected in the absence of EPO (right, n = 5 independent experiments). (B) Arginine uptake in the absence or presence of rEPO (day 4) was monitored using L-[2,3,4-3H)] arginine monohydrochloride (2 μCi) for 10 minutes at room temperature. Uptake in the presence of EPO was arbitrarily set at 1 (means of triplicates in 3 independent experiments). (C) Surface SLC7A1 levels were monitored at days 4, 7, and 10 of rEPO-mediated differentiation and representative histograms are shown (left). Mean fluorescent intensities (MFIs) of SLC7A1 staining relative to day 4 were quantified (right, n = 9). (D) Arginine uptake was evaluated at days 4, 7, and 10 of erythroid differentiation; arginine uptake at day 4 was arbitrarily set at 1 (means of triplicates in 4 independent experiments). (E) CD34+ progenitors were transduced 3 days with GFP-tagged shCTRL and shSLC7A1 lentiviral vectors and representative histograms of cell-surface SLC7A1 expression on GFP+ cells (left) as well as quantification of SLC7A1 expression relative to control-transduced cells is shown (right, n = 8 independent experiments). (F) Arginine uptake was monitored in fluorescence-activated cell sorter–sorted progenitors transduced with shCTRL and shSLC7A1 vectors at day 4 and uptake levels relative to the shCTRL condition are presented (n = 3). (G) The evolution of shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of GFP expression at days 0, 3, and 7 of rEPO-induced differentiation and representative histograms are shown (left). Quantification of the percentages of GFP+ cells relative to day 0 is presented (right, n = 6). (H) Differentiation of rEPO-induced shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of CD34 and CD36 expression on IL3R−GlyA− cells; CFU-E are defined by an IL3R−GlyA−CD34−CD36+ phenotype (left, day 3). Quantification of CFU-E relative to shCTRL-transduced progenitors is presented (right, n = 9). (I) The differentiation of progenitors to an erythroid (GlyA) vs myeloid (CD11b) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative plots (left) and quantifications (right) are presented. Surface expression of GlyA and CD11b was monitored on shCTRL- and shSLC7A1-transduced progenitors at day 3 of differentiation (left). Quantification of erythroid (n = 14) and myeloid (n = 12) differentiation is shown in independent experiments (right). (J) CD34+CD38− progenitors transduced with shCTRL and shCAT1 vectors were evaluated for CFU potential using the StemMACS HSC-CFU Assay Kit and the numbers of burst-forming unit erythroid (BFU-E) and CFU–granulocyte-macrophage (GM) colonies generated at day 14 are presented for 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. IgG, immunoglobulin G; ns, not significant.
To assess whether SLC7A1-mediated arginine metabolism plays a key role in erythropoiesis, we downregulated SLC7A1 expression in cytokine-stimulated progenitors with a pre-erythroid progenitor phenotype,3 characterized as CD34+IL3RαLoCD36− (supplemental Figure 1F). Progenitors transduced with an SLC7A1 shRNA lentiviral vector, also harboring the green fluorescent protein (GFP) reporter gene, exhibited significantly reduced SLC7A1 transcripts (P < .05) (supplemental Figure 1G) and cell-surface SLC7A1 levels (P < .001) (Figure 1E), compared with HSPCs transduced with a control (shCTRL) vector. SLC7A1 downregulation decreased expansion by twofold (P < .0001) (supplemental Figure 1H) but more importantly, these cells were negatively selected; shSLC7A1-transduced progenitors (GFP+) decreased from 54% to 9% in a representative experiment (P < .001) (Figure 1G). Importantly, these progenitors were not negatively selected after expansion in nonerythroid conditions (supplemental Figure 1I). Consistent with an important function for SLC7A1 in erythroid differentiation, its downregulation resulted in the reduction of the IL3R−GlyA−CD34−CD36+ colony-forming unit (CFU)–enriched subset (P < .01) (Figure 1H) and decreased the expression of GlyA, CD36, and CD71 erythroid markers (P < .0001) (Figure 1I; supplemental Figure 1J). Notably though, these EPO-signaled HSPCs maintained their myeloid lineage potential, with more than twofold increase in the differentiation of CD11b+ myeloid cells (P < .01) (Figure 1I).
We next evaluated the impact of SLC7A1 downregulation on the erythroid specification of primitive CD34+CD38− progenitors. After transduction with an shCTRL or shSLC7A1/CAT1 vector, fluorescence-activated cell sorter–sorted GFP+CD34+CD38− bone marrow (BM) progenitors (supplemental Figure 2A) were evaluated in a single-cell StemMACS HSC-CFU Assay Kit (Miltenyi). Assessment of burst-forming unit erythroid progenitors revealed a more than threefold decrease in the number of colonies generated after SLC7A1/CAT1 downregulation (P < .05) (Figure 1J; supplemental Figure 2B). However, the generation of CFU–granulocyte-macrophage, albeit lower, was not significantly attenuated as compared with control conditions (Figure 1J). Furthermore, GFP+CD34+CD38− BM progenitors evaluated in EPO-stimulated liquid culture conditions exhibited attenuated induction of GlyA and only minimal expansion of shCAT1-transduced cells after 7 days (compared with a ninefold expansion of shCTRL-transduced cells, supplemental Figure 2C). These data correlated with a reduced growth of single-sorted GFP+CD34+CD38− cord blood progenitors after shSLC7A1/CAT1 transduction, with growth in 24 of 177 wells as compared with 81 of 177 wells, respectively. Micrographs of wells seeded with 100 sorted GFP+CD34+CD38− shCTRL-transduced cord blood progenitors showed a larger area than after shSLC7A1/CAT1 transduction and a loss of transgene (shSLC7A1 with GFP) in GlyA+ erythroblasts generated from 2 of the 3 wells, attesting to the importance of this transporter (supplemental Figure 2D). Together, these data strongly suggest that SLC7A1 plays a nonredundant role in regulating the erythroid differentiation of human HSPCs.
Arginine metabolism is required for erythroid lineage commitment as well as terminal erythroid differentiation
To specifically evaluate the role of arginine in the commitment of HSPCs to an erythroid lineage fate, progenitors were stimulated with rEPO in media depleted of arginine. In arginine-depleted media, a significantly lower percentage of progenitors adopted a CFU-E fate (P < .0001) (Figure 2A), and these cells exhibited a significantly decreased proliferation capacity (supplemental Figure 3A). Moreover, under conditions of arginine depletion, burst-forming unit erythroid progenitors–enriched CD34+CD36− cells (supplemental Figure 3B) exhibited a reduced differentiation toward a CD34−CD36+ CFU-E stage (supplemental Figure 3B). In agreement with the SLC7A1 knockdown experiments (Figure 1), arginine depletion inhibited erythroid differentiation at both days 3 and 7 of rEPO stimulation (P < .05) (Figure 2B; supplemental Figure 3C). These data highlight the critical role of arginine in promoting the commitment of progenitors to an erythroid fate.
Arginine is required for erythroid lineage commitment and its absence attenuates terminal differentiation. (A) The fate of progenitors treated with rEPO in the presence (+) or absence (−) or exogenous arginine was monitored as a function of CD34 and CD36 expression in IL3R−GlyA− cells at day 3 of differentiation. Representative dot plots are presented (top) and IL3R−GlyA−CD34−CD36+ (CFU-E) cells were quantified relative to differentiation in the presence of arginine (bottom, n = 13). (B) Relative levels of erythroid and myeloid differentiation were monitored by GlyA and CD11b staining, respectively. Representative dot plots are shown at days 3 and 7 of differentiation in the presence or absence of arginine (top). Quantification of GlyA+ (n = 17, day 3; n = 9, day 7) and CD11b+ cells (n = 16, day 3; n = 7, day 7) are presented (bottom). (C) The impact of arginine at later stages of erythroid differentiation was evaluated by depleting arginine after 3 days of EPO-induced erythroid differentiation. GlyA was evaluated at day 3 (top histogram; gray histograms, isotype control; solid line histograms, specific staining) and then 4 days later (day 7) in the presence or absence of arginine (bottom histograms, solid line, and orange histograms, respectively). Quantification of the percentages of GlyA+ cells was compared after EPO-induced differentiation from day 3 to 7 in the presence or absence of arginine (right, n = 9). (D) The impact of arginine deprivation between days 3 and 10 of rEPO-induced differentiation was monitored as a function of CD49d/GLUT1 profiles (top) and enucleation (bottom, Syto16 staining). Representative plots are shown (left) and quantification in 8 individual donors is presented (right). (E) Arginase 2 expression was evaluated by immunoblot at days 4 and 7 of differentiation in the absence or presence of EPO. Actin levels are shown as a loading control (left). Quantification of arginase 2 expression relative to actin was evaluated in the different conditions (right, n = 3). ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
Arginine is required for erythroid lineage commitment and its absence attenuates terminal differentiation. (A) The fate of progenitors treated with rEPO in the presence (+) or absence (−) or exogenous arginine was monitored as a function of CD34 and CD36 expression in IL3R−GlyA− cells at day 3 of differentiation. Representative dot plots are presented (top) and IL3R−GlyA−CD34−CD36+ (CFU-E) cells were quantified relative to differentiation in the presence of arginine (bottom, n = 13). (B) Relative levels of erythroid and myeloid differentiation were monitored by GlyA and CD11b staining, respectively. Representative dot plots are shown at days 3 and 7 of differentiation in the presence or absence of arginine (top). Quantification of GlyA+ (n = 17, day 3; n = 9, day 7) and CD11b+ cells (n = 16, day 3; n = 7, day 7) are presented (bottom). (C) The impact of arginine at later stages of erythroid differentiation was evaluated by depleting arginine after 3 days of EPO-induced erythroid differentiation. GlyA was evaluated at day 3 (top histogram; gray histograms, isotype control; solid line histograms, specific staining) and then 4 days later (day 7) in the presence or absence of arginine (bottom histograms, solid line, and orange histograms, respectively). Quantification of the percentages of GlyA+ cells was compared after EPO-induced differentiation from day 3 to 7 in the presence or absence of arginine (right, n = 9). (D) The impact of arginine deprivation between days 3 and 10 of rEPO-induced differentiation was monitored as a function of CD49d/GLUT1 profiles (top) and enucleation (bottom, Syto16 staining). Representative plots are shown (left) and quantification in 8 individual donors is presented (right). (E) Arginase 2 expression was evaluated by immunoblot at days 4 and 7 of differentiation in the absence or presence of EPO. Actin levels are shown as a loading control (left). Quantification of arginase 2 expression relative to actin was evaluated in the different conditions (right, n = 3). ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
Because SLC7A1 levels decreased during terminal erythropoiesis (Figure 1C), we assessed whether arginine was required during this stage of differentiation. When arginine was depleted from erythroblasts at day 3 of differentiation, all cells upregulated GlyA by day 7 (P < .05) (Figure 2C), albeit with a decrease in cell expansion (P < .001) (supplemental Figure 3D). Notably though, terminal differentiation as well as enucleation was markedly inhibited (P < .01) (Figure 2D). These data were somewhat surprising given the loss in cell-surface SLC7A1 levels upon terminal differentiation (Figure 1C). We therefore evaluated the possibility that intracellular arginine was regulated by arginase expression. Importantly, Arg2 was upregulated by 12-fold during erythroid differentiation and remained elevated in late-stage erythroblasts (P < .01) (Figure 2E), whereas Arg1 was upregulated at very late-stage terminal differentiation (supplemental Figure 3E). Thus, the strong induction of these arginases may be associated with the continued requirement for extracellular arginine throughout the erythroid differentiation process.
Erythroid lineage specification is dependent on arginine-dependent polyamine biosynthesis
Arginine is a precursor of numerous metabolic intermediates that are generated through distinct catabolic pathways (supplemental Figure 3F). Because neither arginine-derived synthesis of creatine nor nitric oxide were required for erythropoiesis (supplemental Figure 3G), we evaluated the role of arginine as a precursor of polyamines (Figure 3A). Notably, polyamine biosynthesis was strictly required for erythroid differentiation: inhibition of polyamine biosynthesis, via the targeting of ornithine carboxylase or spermidine synthase with α-difluoromethylornithine (DFMO) and trans-4-methyl cyclohexylamine (MCHA), respectively, inhibited expansion and abrogated erythroid differentiation (P < .001 to P < .0001) (Figure 3B-C; supplemental Figure 4A-C). In accord with the arginine deprivation experiments, progenitors treated with either DFMO or MCHA maintained their potential to differentiate to a myeloid cell fate, with up to 80% CD11b+ cells (Figure 3B-C). Similarly, activation of SAT1-mediated polyamine catabolism by N1, N11-diethylnorspermine decreased GlyA (P < .0001) while supporting CD11b induction (P < .05) (Figure 3D). Spermidine, and not spermine, was specifically required for erythropoiesis, as inhibiting spermine synthase with N-(3-amino-propyl)cyclohexylamine did not negatively affect erythroid differentiation (Figure 3E). Furthermore, in contrast with the requirement of arginine for both erythroid commitment and terminal erythroid differentiation, polyamine biosynthesis was a prerequisite for erythroid commitment but not for terminal erythroid differentiation. DFMO inhibited the generation of erythroblasts but did not modulate either erythroblast differentiation or enucleation (supplemental Figure 4D-E). Furthermore, enucleation was actually inhibited by spermidine (supplemental Figure 4F), likely because of spermidine’s potential to support mitochondrial respiration, a metabolic state that negatively affects enucleation.14,30
Erythroid differentiation is dependent on arginine-derived polyamine biosynthesis but not transport. (A) Schematic representation of polyamine biosynthesis from arginine with key enzymes indicated in red (ornithine carboxylase [ODC], spermidine synthase [SRM], spermine synthase [SMS], spermidine/spermine-N1-acetyl transferase [SAT1]). Steps that are inhibited by α-difluoromethylornithine (DMFO), trans-4-methyl cyclohexylamine (MCHA), and N-(3-amino-propyl)cyclohexylamine (APCHA) are presented. N1, N11-diethylnorspermine (DENS), an activator of polyamine catabolism, is also indicated. (B) The impact of DFMO (1 mM) on rEPO-induced differentiation of CD34+ progenitors was monitored by evaluating CD11b/GlyA profiles at days 3 and 7. Representative dot plots (top) and quantification of relative levels at days 3 (n = 14 for GlyA, n = 12 for CD11b) and 7 (n = 11 for GlyA, n = 9 for CD11b) of differentiation are shown (bottom). (C) The impact of MCHA (100 μM) was evaluated as a function of CD11b/GlyA profiles (top) at day 3 and quantifications are shown (bottom, n = 7). (D) The impact of DENS (10 μM) on EPO-induced differentiation was evaluated at day 3 and representative plots (top) and quantifications (n = 7 for GlyA and n = 5 for CD11b) are shown (bottom). (E) APCHA (100 μM) was added to rEPO-induced progenitors and representative profiles (top) and quantifications (bottom, n = 7) are shown. (F) DFMO-treated progenitors were differentiated with rEPO in the presence or absence of putrescine (Put; 100 μM) or spermidine (Spd; 100 μM), and GlyA was evaluated at day 7 (top, green histograms). Quantification from 7 independent experiments is presented (bottom). (G) Erythroid differentiation was induced in the presence or absence of DFMO, spermidine, and AMXT-1501 (2.5 μM), an inhibitor of polyamine transport. Quantification of GlyA expression relative to control conditions is presented (bottom, n = 3). ∗P < .05; ∗∗P < .001; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Erythroid differentiation is dependent on arginine-derived polyamine biosynthesis but not transport. (A) Schematic representation of polyamine biosynthesis from arginine with key enzymes indicated in red (ornithine carboxylase [ODC], spermidine synthase [SRM], spermine synthase [SMS], spermidine/spermine-N1-acetyl transferase [SAT1]). Steps that are inhibited by α-difluoromethylornithine (DMFO), trans-4-methyl cyclohexylamine (MCHA), and N-(3-amino-propyl)cyclohexylamine (APCHA) are presented. N1, N11-diethylnorspermine (DENS), an activator of polyamine catabolism, is also indicated. (B) The impact of DFMO (1 mM) on rEPO-induced differentiation of CD34+ progenitors was monitored by evaluating CD11b/GlyA profiles at days 3 and 7. Representative dot plots (top) and quantification of relative levels at days 3 (n = 14 for GlyA, n = 12 for CD11b) and 7 (n = 11 for GlyA, n = 9 for CD11b) of differentiation are shown (bottom). (C) The impact of MCHA (100 μM) was evaluated as a function of CD11b/GlyA profiles (top) at day 3 and quantifications are shown (bottom, n = 7). (D) The impact of DENS (10 μM) on EPO-induced differentiation was evaluated at day 3 and representative plots (top) and quantifications (n = 7 for GlyA and n = 5 for CD11b) are shown (bottom). (E) APCHA (100 μM) was added to rEPO-induced progenitors and representative profiles (top) and quantifications (bottom, n = 7) are shown. (F) DFMO-treated progenitors were differentiated with rEPO in the presence or absence of putrescine (Put; 100 μM) or spermidine (Spd; 100 μM), and GlyA was evaluated at day 7 (top, green histograms). Quantification from 7 independent experiments is presented (bottom). (G) Erythroid differentiation was induced in the presence or absence of DFMO, spermidine, and AMXT-1501 (2.5 μM), an inhibitor of polyamine transport. Quantification of GlyA expression relative to control conditions is presented (bottom, n = 3). ∗P < .05; ∗∗P < .001; ∗∗∗P < .001; ∗∗∗∗P < .0001.
To assure that the impact of pharmacological inhibitors was directly coupled to polyamine biosynthesis, we performed rescue experiments and found that ectopic putrescine and spermidine restored the DFMO- and MCHA-mediated attenuation of erythroid differentiation (P < .0001) (Figure 3F; supplemental Figure 4G). Interestingly, under control conditions, erythroid differentiation was not dependent on polyamine transport from extracellular stores (Figure 3G). However, after DFMO-mediated abrogation of polyamine biosynthesis, treatment with the AMXT-1501 polyamine transporter inhibitor31 abrogated the potential of spermidine to rescue erythropoiesis (Figure 3G; supplemental Figure 4H). These data point to an evolutionary redundancy; intracellular polyamine synthesis is sufficient for erythroid differentiation under physiological conditions, but under stress conditions where intracellular polyamine levels are limiting, the transport of extracellular polyamines promotes erythropoiesis.
The DHPS-catalyzed generation of hypusine from spermidine is required for erythroid differentiation
The pleiotropic nature of polyamines has made it difficult to define their specific role(s).32 Notably though, spermidine serves as a precursor for the generation of the natural amino acid hypusine [Nε-4-amino-2-hydroxybutyl(lysine)], an essential posttranslational modification of eIF5A. Hypusine is formed by the conjugation of the aminobutyl moiety of spermidine to lysine-50 of human eIF5A.33 Remarkably, eIF5A is the only protein in which this modification is known to occur, and its presence is required for its function, promoting translation elongation as well as translation termination.19-21 The synthesis of hypusine is catalyzed through sequential enzymatic steps involving DHPS and deoxyhypusine hydroxylase (DOHH) (Figure 4A).
DHPS is required for erythroid commitment and differentiation. (A) Schematic representation of the 2-step process resulting in eIF5AH. In the first step, DHPS catalyzes the addition of an aminobutyl moiety from the spermidine to the lysine 50 on eIF5A, generating an eIF5A intermediate. Subsequently, DOHH catalyzes the hydroxylation of the spermidine modification, generating an active hypusinated eIF5A. GC7 can inhibit the first step of this reaction. (B) The effect of GC7 on early erythropoiesis was evaluated by monitoring CD34/CD36 profiles of EPO-stimulated CD34+ progenitors in the absence (−) or presence (+) of GC7 (5 μM). Representative dot plots of IL3R−GlyA− cells are shown and the percentages of IL3R−GlyA−CD34−CD36+ (CFU-E) are indicated (left). Quantification of CFU-E in 16 independent experiments are presented (right). (C) Representative dot plots of GlyA/CD11b profiles are presented at days 3 and 7 of EPO-induced differentiation in the absence or presence of GC7 (top). Quantification of GlyA+ and CD11b+ cells relative to levels in the absence of GC7 (indicated as “1”) are presented (bottom, n = 11-19). (D) CD34+ progenitors were differentiated in rEPO for 3 days and differentiation continued until day 7 in the absence or presence of GC7 (between days 3 and 7). Representative histograms showing GlyA expression at days 3 and 7 (with isotype controls, gray histograms) are presented and quantification of GlyA+ cells are presented relative to levels in the absence of GC7 (designated as “1,” bottom, n = 8). (E) CD34+ progenitors were transduced with shCTRL or shDHPS lentiviral vectors, harboring the enhanced GFP transgene and rEPO added 72 hours later. GFP expression was monitored at this time point (designated day 0), as well at days 3 and 7 of differentiation and representative are shown (top). Quantification of the evolution of GFP expression relative to day 0 (designated as “100%”) is presented for 5 donors (bottom). (F) shCTRL and shDHPS transduced progenitors were differentiated in the presence of rEPO for 3 days and representative CD34/CD36 profiles of IL3R−GlyA− cells are presented (top). Quantification of IL3R−GlyA−CD34−CD36+ cells in shDHPS-transduced cells relative to shCTRL-transduced cells are shown (bottom, n = 9). (G) shCTRL and shDHPS transduced progenitors were differentiated for 3 days and representative GlyA/CD11 dot plots are shown (top). Quantification of GlyA+ and CD11b+ cells are presented relative to shCTRL conditions (bottom; n = 20 for GlyA, n = 21 for CD11b). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
DHPS is required for erythroid commitment and differentiation. (A) Schematic representation of the 2-step process resulting in eIF5AH. In the first step, DHPS catalyzes the addition of an aminobutyl moiety from the spermidine to the lysine 50 on eIF5A, generating an eIF5A intermediate. Subsequently, DOHH catalyzes the hydroxylation of the spermidine modification, generating an active hypusinated eIF5A. GC7 can inhibit the first step of this reaction. (B) The effect of GC7 on early erythropoiesis was evaluated by monitoring CD34/CD36 profiles of EPO-stimulated CD34+ progenitors in the absence (−) or presence (+) of GC7 (5 μM). Representative dot plots of IL3R−GlyA− cells are shown and the percentages of IL3R−GlyA−CD34−CD36+ (CFU-E) are indicated (left). Quantification of CFU-E in 16 independent experiments are presented (right). (C) Representative dot plots of GlyA/CD11b profiles are presented at days 3 and 7 of EPO-induced differentiation in the absence or presence of GC7 (top). Quantification of GlyA+ and CD11b+ cells relative to levels in the absence of GC7 (indicated as “1”) are presented (bottom, n = 11-19). (D) CD34+ progenitors were differentiated in rEPO for 3 days and differentiation continued until day 7 in the absence or presence of GC7 (between days 3 and 7). Representative histograms showing GlyA expression at days 3 and 7 (with isotype controls, gray histograms) are presented and quantification of GlyA+ cells are presented relative to levels in the absence of GC7 (designated as “1,” bottom, n = 8). (E) CD34+ progenitors were transduced with shCTRL or shDHPS lentiviral vectors, harboring the enhanced GFP transgene and rEPO added 72 hours later. GFP expression was monitored at this time point (designated day 0), as well at days 3 and 7 of differentiation and representative are shown (top). Quantification of the evolution of GFP expression relative to day 0 (designated as “100%”) is presented for 5 donors (bottom). (F) shCTRL and shDHPS transduced progenitors were differentiated in the presence of rEPO for 3 days and representative CD34/CD36 profiles of IL3R−GlyA− cells are presented (top). Quantification of IL3R−GlyA−CD34−CD36+ cells in shDHPS-transduced cells relative to shCTRL-transduced cells are shown (bottom, n = 9). (G) shCTRL and shDHPS transduced progenitors were differentiated for 3 days and representative GlyA/CD11 dot plots are shown (top). Quantification of GlyA+ and CD11b+ cells are presented relative to shCTRL conditions (bottom; n = 20 for GlyA, n = 21 for CD11b). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
The addition of the spermidine analog GC7, one of the most potent inhibitors of DHPS34 (Figure 4A), blocked the EPO-induced generation of CFU-E (P < .0001) (Figure 4B). Furthermore, myeloid differentiation occurred despite a decrease in progenitor expansion in this condition (P < .0001) (supplemental Figure 5A). CD11b+ cells were increased by 30-fold by day 7 of EPO-induced differentiation (P < .05) (Figure 4C). This increase was accompanied by a massive loss of erythroid progenitors (P < .0001) (Figure 4C-D; supplemental Figure 5B-C). Furthermore, inhibition of DHPS activity at day 3 attenuated the differentiation of late-stage CD49d− orthochromatic erythroblasts and enucleation (P < .01) (supplemental Figure 5D). A pharmacological inhibitor of DOHH, the iron chelator ciclopirox olamine (supplemental Figure 5E),35 also significantly inhibited the induction of GlyA to levels <10% of that detected in control conditions (P < .001) (supplemental Figure 5F).
To directly evaluate hypusination, we targeted DHPS using an shRNA-mediated approach. shDHPS-transduced progenitors exhibited a 61% ± 3% decrease in DHPS messenger RNA (P < .01) (supplemental Figure 5G) and a decreased expansion (P < .0001) (supplemental Figure 5H). Moreover, progenitors with downregulated DHPS were massively counterselected during erythroid differentiation (P < .001) (Figure 4E). Similarly to the impact of GC7, shDHPS-transduced progenitors exhibited a decreased generation of CFU-E (P < .01) (Figure 4F) and a significantly attenuated upregulation of erythroid markers (P < .0001) (Figure 4G). The negative impact of DHPS downregulation was also restricted to erythroid-committed progenitors; in the absence of rEPO, shDHPS-transduced progenitors were not negatively selected (supplemental Figure 5I), and there was a threefold expansion of myeloid cells (P < .001) (Figure 4G). Thus, in a manner analogous to conditions of decreased arginine uptake, inhibition of DHPS-attenuated erythroid but not myeloid differentiation.
Hypusination of eIF5A in erythroid progenitors regulates the translation of proteins involved in cell cycle, protein metabolism, and mitochondrial translation
The data presented above strongly suggest a critical role for hypusination in erythroid differentiation. However, it is notable that eIF5AH has not previously been evaluated as a function of erythroid differentiation. We found that eIF5A levels increased within 4 days of EPO stimulation (P < .01) (Figure 5A) and then dropped during terminal differentiation between polychromatic and orthochromatic erythroblasts (P < .05) (Figure 5B; supplemental Figure 6A).
DHPS-mediated hypusination of eIF5A regulates protein synthesis in early erythroid progenitors. (A) Hypusination was evaluated in progenitors differentiated in the presence or absence of EPO (day 4) and representative immunoblots of eIF5AH, eIF5A, and actin are shown (left). Quantification of eIF5A/actin (middle) and eIF5AH/eIF5A (right) ratios are presented relative to levels in the presence of EPO (n = 7). (B) Proerythroblasts (Pro), basophilic (Baso), polychromatic (Poly), and orthochromatic (Ortho) erythroblasts were sorted based on their GLUT1/CD49d prolife at day 7 of differentiation and hypusination was monitored by immunoblotting (left). eIF5AH was quantified relative to total eIF5A levels (right, n = 5). (C) Protein synthesis was monitored at the indicated day of erythroid differentiation by O-propargyl-puromycin (OPP) labeling and representative histograms and MFI are indicated (left). Quantification of MFIs relative to day 4 are presented for 3 donors (right). (D) Protein synthesis was monitored by OPP labeling in erythroblast subsets 24 hours after sorting (as in panel B, left). Quantification of MFIs relative to proerythroblasts are presented for 3 donors (right). (E) Hypusination was evaluated after 3 days of EPO stimulation in the absence (−) or presence (+) of GC7 (5 μM) and representative immunoblots of eIF5AH, eIF5A, and actin are shown (left). Quantification relative to levels in the absence of GC7 are presented (right, n = 9). (F) Hypusination in shCTRL- and shDHPS-transduced progenitorsevaluated at day 3 of differentiation after sorting based on GFP expression and representative immunoblots are shown (left). Quantification of the relative levels of eIF5AH/eIF5A is presented (right, n = 5). (G) CD34+ progenitors were differentiated in the presence of EPO, together with GC7 (5 μM) or cycloheximide (CHX; 1 μM). Protein synthesis was evaluated at day 1 of differentiation and a representative histogram is shown (left). Quantification of protein synthesis relative to control conditions is presented (right, n = 8). (H) shCTRL- and shDHPS-transduced progenitors were sorted 72 hours after transduction based on GFP expression. Protein synthesis evaluated 24 hours after the addition of EPO and a representative histogram (left) as well as quantification (right, n = 3) are presented. (I) CD34+ progenitors were differentiated with EPO in the absence or presence of GC7 (5 μM) for 2 days and protein expression was evaluated by mass spectrometry–based quantitative proteomics. A volcano plot shows differences in protein expression (log2 fold change) induced by GC7, and the identity of specified downregulated and upregulated proteins are noted. Statistical significance of relative protein expression is computed via 2-sample moderated t test, and proteins with an false discovery rate (FDR) adjusted (adj.) P < .05 are colored in red. (J) Overrepresentation analyses of gene ontology for nonredundant biological processes were evaluated for significantly upregulated and downregulated and enrichment scores are indicated. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
DHPS-mediated hypusination of eIF5A regulates protein synthesis in early erythroid progenitors. (A) Hypusination was evaluated in progenitors differentiated in the presence or absence of EPO (day 4) and representative immunoblots of eIF5AH, eIF5A, and actin are shown (left). Quantification of eIF5A/actin (middle) and eIF5AH/eIF5A (right) ratios are presented relative to levels in the presence of EPO (n = 7). (B) Proerythroblasts (Pro), basophilic (Baso), polychromatic (Poly), and orthochromatic (Ortho) erythroblasts were sorted based on their GLUT1/CD49d prolife at day 7 of differentiation and hypusination was monitored by immunoblotting (left). eIF5AH was quantified relative to total eIF5A levels (right, n = 5). (C) Protein synthesis was monitored at the indicated day of erythroid differentiation by O-propargyl-puromycin (OPP) labeling and representative histograms and MFI are indicated (left). Quantification of MFIs relative to day 4 are presented for 3 donors (right). (D) Protein synthesis was monitored by OPP labeling in erythroblast subsets 24 hours after sorting (as in panel B, left). Quantification of MFIs relative to proerythroblasts are presented for 3 donors (right). (E) Hypusination was evaluated after 3 days of EPO stimulation in the absence (−) or presence (+) of GC7 (5 μM) and representative immunoblots of eIF5AH, eIF5A, and actin are shown (left). Quantification relative to levels in the absence of GC7 are presented (right, n = 9). (F) Hypusination in shCTRL- and shDHPS-transduced progenitorsevaluated at day 3 of differentiation after sorting based on GFP expression and representative immunoblots are shown (left). Quantification of the relative levels of eIF5AH/eIF5A is presented (right, n = 5). (G) CD34+ progenitors were differentiated in the presence of EPO, together with GC7 (5 μM) or cycloheximide (CHX; 1 μM). Protein synthesis was evaluated at day 1 of differentiation and a representative histogram is shown (left). Quantification of protein synthesis relative to control conditions is presented (right, n = 8). (H) shCTRL- and shDHPS-transduced progenitors were sorted 72 hours after transduction based on GFP expression. Protein synthesis evaluated 24 hours after the addition of EPO and a representative histogram (left) as well as quantification (right, n = 3) are presented. (I) CD34+ progenitors were differentiated with EPO in the absence or presence of GC7 (5 μM) for 2 days and protein expression was evaluated by mass spectrometry–based quantitative proteomics. A volcano plot shows differences in protein expression (log2 fold change) induced by GC7, and the identity of specified downregulated and upregulated proteins are noted. Statistical significance of relative protein expression is computed via 2-sample moderated t test, and proteins with an false discovery rate (FDR) adjusted (adj.) P < .05 are colored in red. (J) Overrepresentation analyses of gene ontology for nonredundant biological processes were evaluated for significantly upregulated and downregulated and enrichment scores are indicated. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
The potential impact of hypusination on protein synthesis was evaluated as a function of the incorporation of the alkyne analog of puromycin, O-propargyl-puromycin, forming covalent conjugates with nascent polypeptide chains.36,37 Within 1 day of EPO stimulation, O-propargyl-puromycin incorporation was significantly increased, indicating an augmented level of protein synthesis (P < .05) (supplemental Figure 6B), and then decreased during terminal differentiation (Figure 5C-D). Interestingly, the dramatic drop in protein synthesis occurred earlier than the drop in hypusination, between the basophilic and polychromatic erythroblast stages (P < .001) (Figure 5D). These data correlate with previous findings showing that total protein levels also drop between the basophilic and polychromatic stages of differentiation.38
To assess whether hypusination in hematopoietic progenitors is directly regulated by arginine catabolism, we evaluated hypusination after arginine deprivation and polyamine biosynthesis inhibition. Both these conditions dramatically reduced hypusination without having a significant impact on eIF5A levels themselves (supplemental Figure 6C-D). Furthermore, polyamine biosynthesis was directly required for hypusination, and putrescine and spermidine rescued the DFMO-mediated inhibition of hypusination (P < .001) (supplemental Figure 6D). Hypusination also directly affected the activation state of the mTOR pathway, integrating environmental clues into cellular responses. Phosphorylation of RPS6, a downstream reporter of mTOR activity, was inhibited by GC7 and DFMO to levels comparable with those detected after arginine deprivation (supplemental Figure 6E). Thus, hypusination in hematopoietic progenitors is strictly dependent on arginine-derived polyamine biosynthesis and regulates mTOR signaling cascades.
As expected from these results, inhibition of DHPS, by either GC7 or an shDHPS, resulted in a 75% loss of hypusine as compared with that in control progenitors (P < .0001 and P < .01) (Figure 5E-F). Moreover, these modulations resulted in functional changes in protein synthesis. Nascent protein synthesis in GC7-treated progenitors decreased to levels detected in cells treated with a global inhibitor of protein synthesis (P < .0001) (Figure 5G-H).
To specifically evaluate the proteins that were regulated by hypusinated eIF5A in erythroid progenitors, we analyzed progenitors differentiated for 48 hours in the absence or presence of GC7 by quantitative liquid chromatography tandem mass spectrometry–based proteomics using tandem mass tags.39 Notably, and in agreement with the impact of GC7 on erythroid differentiation, some of the most highly downregulated proteins were hemoglobin subunits and erythroid spectrin chains (Figure 5I). Furthermore, DHPS was significantly upregulated after GC7 treatment, pointing to a potential compensatory feedback loop.
Gene set enrichment analysis and protein-protein interaction network clustering analysis revealed a massive loss of proteins involved in the translation of mitochondrial proteins (Figure 5J; supplemental Figure 7A). Our proteomics analyses identified 824 of 1158 mitochondrial proteins; after attenuation of eIF5AH, 620 proteins were downregulated and 204 were upregulated (supplemental Figure 7B), with a dramatic decrease in proteins involved in oxidative phosphorylation (supplemental Figure 7C). Importantly, eIF5A functions in cytoplasmic, not mitochondrial, protein synthesis, and thus, the impact on mitochondrial proteins may be either direct or indirect.40 Although the mechanisms regulating the massive impact of hypusination on mitochondrial proteins is not known, this effect has been detected in multiple other cell systems, including renal cells,41 brain,42 hepatic cells,43 cardiomyocytes,44 macrophages,45 and T cells.46 In the context of red cells, these data are of much interest given previous work showing the importance of mitochondrial reactive oxygen species in erythroid differentiation.14,30,47 Indeed, we found that the specific inhibition of mitochondrial protein synthesis with chloramphenicol significantly decreased erythroid progenitor differentiation (P < .001), whereas CD11b+ myeloid cells continued to be generated (P < .05) (supplemental Figure 7D). Thus, although there is a large literature reporting on the importance of protein synthesis during erythropoiesis,22,48,49 our results highlight the critical role of cytoplasmic hypusine-dependent protein synthesis in the mitochondrial translation apparatus required for erythroid differentiation.
The attenuated mitochondrial metabolism and erythroid differentiation associated with defective hypusination are partially rescued by succinate
To decipher the impact of hypusination on the synthesis of proteins involved in mitochondrial metabolism, we first evaluated the expression of mitochondrial complex enzymes. Interestingly, complexes II, III, and V were all upregulated by EPO-induced differentiation (P < .05) (supplemental Figure 8A). Notably though, both GC7 and DHPS downregulation resulted in decreased expression of all 5 mitochondrial complex enzymes, whereas actin levels remained stable (Figure 6A). Thus, the expression levels of this subset of mitochondrial proteins, synthesized in the cytoplasm, are distinctively sensitive to eIF5AH levels.
Hypusination-induced OXPHOS is required for the erythroid commitment of hematopoietic progenitors. (A) Mitochondrial complexes (CI to CV) were monitored on progenitors treated with GC7 or after transduction with shCTRL or shDHPS vectors (day 3) using the OXPHOS monoclonal antibody cocktail (left). Quantification relative to control conditions was determined (n = 4 for GC7 and n = 3 for shRNA-transduced progenitors; middle and right, respectively). (B) Oxygen consumption rate (OCR), a measure of OXPHOS, was monitored on day 1 of erythroid differentiation in the absence or presence of GC7 (5 μM) on a Seahorse XFe96 analyzer after sequential injection of oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and Rotenone/Antimycin A (Rot/AntA; arrows, left). Mean basal OCR and SRC levels ± standard error of the mean (SEM) are presented (middle, n = 4). Representative energy plots of basal OCR and extracellular acidification rate (ECAR), a measure of glycolysis, are presented (right). (C) OCR was monitored on fluorescence-activated cell sorter–sorted shCTRL- and shDHPS-transduced progenitors at day 1 of differentiation (left). Basal OCR and SRC levels ± SEM were evaluated in 4 independent experiments (middle) and a representative OCR/ECAR energy plot is presented (right). (D) OCR was monitored on CD34+ progenitors differentiated with EPO for 24 hours in the absence or presence of GC7 (5 μM) and succinate (SUC, 5 mM) and representative graphs are shown (left). Basal OCR and SRC levels in 6 independent experiments are presented (right). (E) Erythroid differentiation in progenitors treated with EPO in the absence or presence of GC7 or succinate was evaluated at day 7 as a function of GlyA expression and representative histograms are shown (left). Quantification of the percentages of GlyA+ cells relative to control conditions are presented (right, n = 9). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, ∗∗∗∗P < .0001.
Hypusination-induced OXPHOS is required for the erythroid commitment of hematopoietic progenitors. (A) Mitochondrial complexes (CI to CV) were monitored on progenitors treated with GC7 or after transduction with shCTRL or shDHPS vectors (day 3) using the OXPHOS monoclonal antibody cocktail (left). Quantification relative to control conditions was determined (n = 4 for GC7 and n = 3 for shRNA-transduced progenitors; middle and right, respectively). (B) Oxygen consumption rate (OCR), a measure of OXPHOS, was monitored on day 1 of erythroid differentiation in the absence or presence of GC7 (5 μM) on a Seahorse XFe96 analyzer after sequential injection of oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and Rotenone/Antimycin A (Rot/AntA; arrows, left). Mean basal OCR and SRC levels ± standard error of the mean (SEM) are presented (middle, n = 4). Representative energy plots of basal OCR and extracellular acidification rate (ECAR), a measure of glycolysis, are presented (right). (C) OCR was monitored on fluorescence-activated cell sorter–sorted shCTRL- and shDHPS-transduced progenitors at day 1 of differentiation (left). Basal OCR and SRC levels ± SEM were evaluated in 4 independent experiments (middle) and a representative OCR/ECAR energy plot is presented (right). (D) OCR was monitored on CD34+ progenitors differentiated with EPO for 24 hours in the absence or presence of GC7 (5 μM) and succinate (SUC, 5 mM) and representative graphs are shown (left). Basal OCR and SRC levels in 6 independent experiments are presented (right). (E) Erythroid differentiation in progenitors treated with EPO in the absence or presence of GC7 or succinate was evaluated at day 7 as a function of GlyA expression and representative histograms are shown (left). Quantification of the percentages of GlyA+ cells relative to control conditions are presented (right, n = 9). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, ∗∗∗∗P < .0001.
Based on the importance of eIF5AH for the synthesis of mitochondrial proteins, we studied mitochondrial metabolism in these cells. Although GC7 treatment and DHPS downregulation both diminished mitochondrial biomass and polarization potential (P < .01) (supplemental Figure 8B-C), decreases were <1.2-fold. However, the impact on metabolism was much more striking; GC7 almost completely abrogated oxidative phosphorylation (OXPHOS) in these progenitors, and their glycolytic reserve was decreased (Figure 6B; supplemental Figure 8D). Similarly, the inhibition of polyamine biosynthesis by either DFMO or N1, N11-diethylnorspermine decreased basal respiration and spare respiratory capacity by 2.5- to 4.5-fold (supplemental Figure 8E). This loss of OXPHOS, albeit with maintained glycolysis, was also detected in progenitors with downregulated DHPS (Figure 6C; supplemental Figure 8F), again highlighting the critical role of hypusination for mitochondrial metabolism. Ciclopirox, an inhibitor of hypusination and mitochondrial respiration,50 also significantly decreased basal and maximal oxygen consumption (P < .05 and P < .001) (supplemental Figure 8G).
The loss of mitochondrial metabolism under conditions where hypusination was negatively affected led us to test the hypothesis that drivers of the tricarboxylic acid cycle would augment erythroid differentiation. Succinate, by bypassing complex I, can potentially provide a fuel source for compromised mitochondria.51 Furthermore, our protein analyses revealed a significant loss of the complex II SDHB enzyme (Figure 6A), thereby attenuating the generation of succinate. Notably, the addition of succinate to GC7-treated EPO-induced progenitors significantly upregulated OXPHOS (P < .01) (Figure 6D), a response not induced by acetate or α-ketoglutarate (data not shown). Even more importantly, ectopic succinate promoted the erythroid lineage commitment of GC7-treated hematopoietic progenitors (P < .01) (Figure 6E). Together, these data reveal an interplay between hypusination and mitochondrial metabolism in regulating erythroid lineage commitment and differentiation.
Defective erythroid differentiation in RP haploinsufficiency is coupled to an attenuated hypusination
Consistent with its role in translation and binding in the E site of the ribosome, eIF5A interacts with proteins associated with ribosome function, including RPL and RPS proteins.52 Of note, mutations resulting in impaired ribosome biogenesis, encompassing >20 RPs have been found to result in DBA. This rare congenital disease predominantly affects erythroid lineage cells and is characterized by macrocytic anemia and BM failure.53 Furthermore, haploinsufficiency of the RPS14 gene results in macrocytic anemia in del(5q) in patients with MDS.54 However, the potential association between RP function and eIF5AH has not been previously evaluated. Notably, we found that eIF5AH was significantly lower in immortalized haploinsufficient Rps14+/− progenitors55 than in control Mx1Cre+ progenitors (P < .01) (supplemental Figure 9A). Moreover, basal oxygen consumption as well as spare respiratory capacity were significantly lower in Rps14+/−Mx1Cre+ than in Mx1Cre+ progenitors (P < .01) (supplemental Figure 9B), thereby linking RPS14 expression to hypusination and mitochondrial metabolism.
We further assessed hypusination in CD34+ progenitors from del(5q) in patients with MDS. Similar to Rps14-haploinsufficient murine progenitors, progenitors from 3 patients with del(5q)-MDS (supplemental Table 1) exhibited high expression of eIF5A, but hypusination levels were significantly reduced (P < .05) (Figure 7A). Moreover, after rEPO-induced differentiation, eIF5AH decreased in del(5q)-MDS progenitors, accompanied by ineffective erythroid differentiation (Figure 7A; supplemental Figure 9C). This defective hypusination was specific to EPO-stimulated CD34+ progenitors in 2 patients with MDS, where we were able to evaluate hypusination in another cell lineage, activated T cells, which exhibited a more than twofold increase in hypusination (supplemental Figure 9D). Thus, in both murine and human progenitors, the ineffective erythropoiesis linked to RPS14 haploinsufficiency is associated with a loss of hypusinated eIF5A.
Haploinsufficiency in RP genes is coupled to an attenuated hypusination in erythroid progenitors. (A) CD34+ BM progenitors from a healthy control and a patient with del(5q)-MDS were expanded ex vivo. Immunoblots showing hypusinated and total eIF5A levels at days 0 and 3 of EPO-induced differentiation are presented (left). Hypusination in CD34+ BM progenitors from a second patient with del(5q)-MDS was compared with progenitors from a patient with refractory anemia, after a 3-day differentiation in the absence (−) or presence (+) of EPO and blots are shown (middle). Quantification of the ratios of eIF5A/actin and eIF5AH/eIF5A for progenitors from 3 patients with del(5q)-MDS are presented (n = 3). (B) CD34+ BM progenitors from a RPL11-haploinsufficient patient with DBA [c.27_28 delGA (p.N10FS)] were evaluated for hypusinated and total eIF5A levels at isolation (day 0) and then expanded for 4 days followed by 3 days of EPO-induced differentiation (left). The change in eIF5AH/eIF5A after differentiation is shown. Peripheral blood (PB) CD34+ progenitors from a second RPL11-haploinsufficient patient (c.164dup; p.Tyr55Ter) were evaluated after ex vivo differentiation with a 4-day expansion followed by a 3 day EPO stimulation. eIF5AH immunoblots are shown (middle). Quantifications of the ratios of eIF5A/actin and eIF5AH/eIF5A for the 2 patients with DBA and 2 controls at day 3 of EPO-induced differentiation are presented (right). (C) The differentiation of shCTRL- and shRPL11-transduced progenitors to a vs n erythroid (GlyA+) vs myeloid (CD11b+) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative CD11b/GlyA dot plots (left) and quantifications are presented (right, n = 10). (D) Hypusination was evaluated in human progenitors transduced with shCTRL and shRPL11 vectors at day 3 of differentiation. Representative immunoblots of eIF5AH and total eIF5A levels in shCTRL- and shRPL11-transduced progenitors (left) and quantifications ± SEM of eIF5AH/eIF5A and eIF5A/actin are presented (right, n = 6). Levels in control cells are set at “1.” (E) OCRs of shCTRL- and shRPL11-transduced progenitors were evaluated at day 1 of differentiation and representative data (left, n = 3-6 technical replicates) and mean basal OCR levels ± SEM are presented (right, n = 3 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗P < .0001.
Haploinsufficiency in RP genes is coupled to an attenuated hypusination in erythroid progenitors. (A) CD34+ BM progenitors from a healthy control and a patient with del(5q)-MDS were expanded ex vivo. Immunoblots showing hypusinated and total eIF5A levels at days 0 and 3 of EPO-induced differentiation are presented (left). Hypusination in CD34+ BM progenitors from a second patient with del(5q)-MDS was compared with progenitors from a patient with refractory anemia, after a 3-day differentiation in the absence (−) or presence (+) of EPO and blots are shown (middle). Quantification of the ratios of eIF5A/actin and eIF5AH/eIF5A for progenitors from 3 patients with del(5q)-MDS are presented (n = 3). (B) CD34+ BM progenitors from a RPL11-haploinsufficient patient with DBA [c.27_28 delGA (p.N10FS)] were evaluated for hypusinated and total eIF5A levels at isolation (day 0) and then expanded for 4 days followed by 3 days of EPO-induced differentiation (left). The change in eIF5AH/eIF5A after differentiation is shown. Peripheral blood (PB) CD34+ progenitors from a second RPL11-haploinsufficient patient (c.164dup; p.Tyr55Ter) were evaluated after ex vivo differentiation with a 4-day expansion followed by a 3 day EPO stimulation. eIF5AH immunoblots are shown (middle). Quantifications of the ratios of eIF5A/actin and eIF5AH/eIF5A for the 2 patients with DBA and 2 controls at day 3 of EPO-induced differentiation are presented (right). (C) The differentiation of shCTRL- and shRPL11-transduced progenitors to a vs n erythroid (GlyA+) vs myeloid (CD11b+) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative CD11b/GlyA dot plots (left) and quantifications are presented (right, n = 10). (D) Hypusination was evaluated in human progenitors transduced with shCTRL and shRPL11 vectors at day 3 of differentiation. Representative immunoblots of eIF5AH and total eIF5A levels in shCTRL- and shRPL11-transduced progenitors (left) and quantifications ± SEM of eIF5AH/eIF5A and eIF5A/actin are presented (right, n = 6). Levels in control cells are set at “1.” (E) OCRs of shCTRL- and shRPL11-transduced progenitors were evaluated at day 1 of differentiation and representative data (left, n = 3-6 technical replicates) and mean basal OCR levels ± SEM are presented (right, n = 3 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗P < .0001.
To further evaluate the link between RPs and hypusination, we studied eIF5AH in CD34+ progenitors from 2 patients with DBA with RPL11 haploinsufficiency (supplemental Table 1). For 1 patient with DBA, where sufficient numbers of freshly isolated BM CD34+ progenitors were available for evaluation, neither eIF5A nor hypusination were attenuated (Figure 7B). Notably though, after EPO-induced stimulation, hypusination decreased by threefold relative to control CD34+ progenitors, and in a second RPL11-haploinsufficient patient, hypusination as well as eIF5A levels in EPO-stimulated peripheral CD34+ progenitors were more than sixfold lower than those in control (Figure 7B). To assess whether this phenotype would be recapitulated by directly downregulating RPL11, an shRNA against this target was introduced into healthy CD34+ progenitors. As expected, RPL11 downregulation (supplemental Figure 9E) decreased erythroid differentiation, whereas myeloid cells continued to be generated (Figure 7C; supplemental Figure 9F).24,25 Strikingly, the shRNA-mediated downregulation of RPL11 significantly decreased hypusination, despite the maintenance of eIF5A levels (P < .01) (Figure 7D). Moreover, the attenuated hypusination in these progenitors directly correlated with a loss in mitochondrial metabolism (Figure 7E; supplemental Figure 9G). Thus, these data reveal a novel role for RPs in controlling eIF5AH in erythroid progenitors, resulting in a regulated coordination of mitochondrial translation and metabolism with subsequent erythroid differentiation.
Discussion
Translation is a critical component in the regulation of multiple developmental pathways, cell activation, and cancer progression.56-59 Within the broad realm of protein synthesis, hypusinated eIF5A drives translational programs affecting cancer, diabetes, and infectious diseases.20,60-62 Notably though, the impact of eIF5A in erythropoiesis, a process that is particularly sensitive to translational regulation, has not been evaluated. The data that we present here show that optimal protein synthesis in erythroid progenitors is dependent on SLC7A1-mediated arginine uptake, resulting in the hypusination of eIF5A by the arginine-catabolized spermidine moiety. Under conditions where hypusination is suboptimal, hematopoietic progenitors are not capable of undergoing erythroid differentiation but maintain their myeloid lineage fate. Thus, our study reveals a direct link between arginine metabolism and eIF5A-induced protein synthesis in erythroid-myeloid lineage commitment. Furthermore, we uncover a critical role for eIF5A activity in the defective erythropoiesis associated with ribosomopathies.
Hypusinated eIF5A has been identified as required for the translation elongation process rather than the initiation process,63,64 especially important for facilitating the translation of proteins containing polyproline residues.65-67 Notably, eIF5A is the only protein in eukaryotes and archaea to contain hypusine.19,64,68 The downstream effectors via which hypusinated eIF5A regulates cell differentiation are still being elucidated, but recent studies show the importance of hypusinated eIF5A in mitochondrial translation.45,46 The mechanisms via which hypusinated eIF5A stimulates translation of messenger RNAs encoding mitochondrial RPs have not yet been elucidated, but it is intriguing to speculate that in addition to a direct effect, it may modulate expression of chaperones involved in the assembly of mitochondrial proteins. Moreover, eIF5A may interact directly with mitofusin, a protein that is critical in mitochondrial clustering and function.40 Irrespective of the mechanism(s), eIF5A-induced mitochondrial programming reverses B-cell senescence,69 promotes vaccine immunogenicity,70 and protects the brain from premature aging,42,71 among others.
The findings that we present here demonstrate the critical nature of eIF5A-dependent mitochondrial function in regulating the erythroid commitment of HSPCs. Erythroid differentiation requires mitochondrial function11,14,47,72,73 and we now find that in the absence of eIF5A activity, mitochondrial complex enzymes are massively decreased, coupled with an almost complete loss of oxygen consumption. Thus, eIF5A-dependent OXPHOS is a sine qua non for erythroid differentiation. Furthermore, the abrogated erythroid commitment of DHPS-attenuated progenitors can be partially rescued by succinate. Interestingly, although succinate can increase oxygen consumption, its accumulation can modulate cell physiology through succinylation of substrates,74 as well as by generating a condition of “pseudohypoxia” secondary because of stabilization of HIF-α prolyl hydroxylase.75
The rapid initiation of translation allows erythroid progenitors to immediately respond to environmental cues, stimulating erythroid differentiation programs.49,76 It is therefore not surprising that multiple translation initiation factors, such as eIF4G, eIF2α, and EIF4EBP1, have been found to regulate erythropoiesis.77-81 Moreover, haploinsufficiency of RPs affects the translation of specific transcripts,22,23,82-84 resulting in anemia in patients with DBA and del(5q) MDS.85,86 The mechanisms via which RP deficiency regulates hypusine levels are not known, but one hypothesis is that RP-mediated changes in DHPS, DOHH, spermidine, and/or the substrates and cofactors mediating their activities, including oxygen concentration,87 can lead to reduced hypusine levels. Furthermore, eIF2 plays an important role in erythroid differentiation,79 and as colliding ribosomes have recently been shown to trigger eIF2 phosphorylation,88 impaired eIF5A function may cause translating ribosomes to collide on poly-Pro motifs, contributing to the pathophysiology of DBA. More generally, variations in hypusination as a function of cell type/activation highlight the intricate regulation of metabolic and translational pathways. Considering our data, it is opportune to assess the impact of ribosomal availability on eIF5A-dependent translation, bringing a novel perspective to our understanding of the defective protein synthesis characterizing ribosomopathies.
The link that we uncover between RPs and eIF5A activity opens new therapeutic perspectives for patients with ribosomopathies. Notably, L-leucine has recently been found to increase protein synthesis in both DBA and del(5q) MDS erythroblasts, resulting in improved erythroid differentiation23,89 as well as an erythroid response in ∼15% of patients with DBA.90 We hypothesize that the erythroid protein subsets regulated by the leucine-mediated induction of the eIF4F complex91 may differ from those that are dependent on hypusinated eIF5A. It will be important to assess whether the defective hypusination that we detected in RPS14- and RPL11-haploinsufficient progenitors is also a characteristic of patients with RPS19+/− DBA. eIF5AH can potentially be increased by arginine supplementation or more directly by spermidine itself. Spermidine may have a greater impact than arginine when eIF5A activity is limiting; spermidine exhibited therapeutic efficacy in cognition and42,92,93 reversed B-cell senescence69; and improved vaccinal responses in the elderly.70 In erythroid differentiation, results have been disparate,94,95 potentially because of the presence of amine oxidases in fetal calf serum, which catalyze the generation of hyperoxide from spermidine.96 Although a therapeutic strategy designed to deliver high doses of spermidine has not yet been established, a prospective cohort study found a significant correlation between high dietary spermidine intake and cognitive function.93 The data that we present here support the clinical evaluation of combined nutrient supplementation in patients with ribosomopathies, with the goal of promoting eIF4F- as well as eIF5A-dependent protein synthesis.
Acknowledgments
The authors thank all members of their laboratories for discussions and scientific critique. The authors are grateful to Jean-Luc Veyrune and the Unité of Thérapie Cellulaire, as well as the Clémentville Clinic (Montpellier, France), for their generous efforts in providing access to cord blood samples, Sandra Manceau and Pierre Buffet of the GR-Ex for their precious assistance with the GR-Ex clinical protocol and patient collection, Isabelle Marie for her valued help with the Diamond-Blackfan anemia (DBA) registry, Thierry Leblanc and Flore Sicre De Fontbrune for their wonderful help with one of the patients with DBA, and Ludivine David Nguyen for her expertise in the molecular DBA diagnosis. The authors are thankful to Steven A. Carr and Karl Clauser for their insightful input on proteomics analyses.
This work was supported by generous funding from the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK32094) (N.M., S.K., and N.T.), Laboratory of Excellence for Red Cells [(LABEX GR-Ex)-ANR Avenir-11-LABX-0005-02] (P.G.-M., M.S., L.D.C., S.K., and N.T.), NIH, NIDDK R01 DK060581 and R01 DK124906 (R.G.M.), EJPRD/ANR-19-RAR4-0016 and EJP RD COFUND-EJP 825575 (L.D.C.), NIH, National Heart, Lung, and Blood Institute (NHLBI) (HL144436 and HL152099) (L.B.), the FRM, ARC, the French national (ANR) research grants (NutriDiff), the French laboratory consortiums (LABEX) EpiGenMed and GR-Ex, and by intramural NIH funding from the NHLBI (C.E.D.), the Eunice Kennedy Shriver National Institute of Child Health and Human Development grant ZIA HD001010 (T.E.D.), and the National Cancer Institute's Center for Cancer Research (ZIA BC 011924) (N.T.). P.G.-M. was supported by a fellowship from the Clarin-COFUND EU Program (Principado de Asturias, Spain), I.P. was supported by a fellowship from EpiGenMed, and A.J. is supported by a fellowship from the French Ministry of Health.
Authorship
Contribution: P.G.-M., S.K., and N.T. conceived the study; P.G.-M., I.P., M.E.O., A.J., J. Papoin, H.Y., J.G., J. Platon, S.W.S.K., K.L.M., R.G.M., M.P., T.K., M.B.-C., N.P.-S., M.S., T.E.D., N.M., L.D.C., N.D.U., L.B., S.K., and N.T. were involved in study design; K.L.M., S.L., D.J.Y., F.P., A.N., G.C., C.E.D., and L.D.C. participated in the analyses of patients, provided samples, and provided clinical information; P.G.-M., I.P., M.E.O., A.J., J. Papoin, H.Y., J.G., J. Platon, S.W.S.K., K.L.M., M.D., M.P., T.K., S.B., and M.B.-C. performed experiments; all authors participated in data analysis; P.G.-M., S.K., and N.T. wrote the initial manuscript with extensive edits from I.P., M.E.O., M.S., T.E.D., N.M., N.D.U., and L.B.; and K.L.M., V.D., and V.S.Z. provided important critical input.
Conflict-of-interest disclosure: M.S., S.K., and N.T. are inventors on patents describing the use of RBD ligands but N.T. no longer has any patent rights. M.S. is the cofounder of METAFORA-biosystems, a start-up company that focuses on metabolite transporters under physiological and pathological conditions. The remaining authors declare no competing financial interests.
Correspondence: Pedro Gonzalez-Menendez, Departamento de Morfología y Biología Celular, School of Medicine, Universidad de Oviedo, Julián Claveria 6, 33006 Oviedo, Spain; e-mail: gonzalezmpedro@uniovi.es; Sandrina Kinet, Institut de Génétique Moléculaire de Montpellier, CNRS, 1919 Route de Mende, 34090 Montpellier, France; e-mail: kinet@igmm.cnrs.fr; and Naomi Taylor, Pediatric Oncology Branch, National Cancer Institute, National Institutes of Health, Building 10, Room 1W-3940, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: taylorn4@mail.nih.gov.
References
Author notes
∗A.J., J. Papoin, H.Y., J.G., and J. Platon contributed equally to this study.
†S.K. and N.T. are joint senior authors.
The original mass spectra, spectral library, and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000091139/.
Data are available on request from the corresponding authors, Pedro Gonzalez-Menendez (gonzalezmpedro@uniovi.es), Sandrina Kinet (sandrina.kinet@igmm.cnrs.fr), and Naomi Taylor (taylorn4@mail.nih.gov).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![HSC specification to the erythroid lineage is dependent on the expression and function of the SLC7A1 arginine transporter. (A) Cell-surface expression of SLC7A1 was evaluatedafter rEPO-induced erythroid differentiation of CD34+ progenitors (day 4) and representative histograms in the absence or presence of rEPO are presented (left). Relative MFIs of SCL7A1 in rEPO-induced progenitors are compared with levels detected in the absence of EPO (right, n = 5 independent experiments). (B) Arginine uptake in the absence or presence of rEPO (day 4) was monitored using L-[2,3,4-3H)] arginine monohydrochloride (2 μCi) for 10 minutes at room temperature. Uptake in the presence of EPO was arbitrarily set at 1 (means of triplicates in 3 independent experiments). (C) Surface SLC7A1 levels were monitored at days 4, 7, and 10 of rEPO-mediated differentiation and representative histograms are shown (left). Mean fluorescent intensities (MFIs) of SLC7A1 staining relative to day 4 were quantified (right, n = 9). (D) Arginine uptake was evaluated at days 4, 7, and 10 of erythroid differentiation; arginine uptake at day 4 was arbitrarily set at 1 (means of triplicates in 4 independent experiments). (E) CD34+ progenitors were transduced 3 days with GFP-tagged shCTRL and shSLC7A1 lentiviral vectors and representative histograms of cell-surface SLC7A1 expression on GFP+ cells (left) as well as quantification of SLC7A1 expression relative to control-transduced cells is shown (right, n = 8 independent experiments). (F) Arginine uptake was monitored in fluorescence-activated cell sorter–sorted progenitors transduced with shCTRL and shSLC7A1 vectors at day 4 and uptake levels relative to the shCTRL condition are presented (n = 3). (G) The evolution of shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of GFP expression at days 0, 3, and 7 of rEPO-induced differentiation and representative histograms are shown (left). Quantification of the percentages of GFP+ cells relative to day 0 is presented (right, n = 6). (H) Differentiation of rEPO-induced shCTRL- and shSLC7A1-transduced progenitors was monitored as a function of CD34 and CD36 expression on IL3R−GlyA− cells; CFU-E are defined by an IL3R−GlyA−CD34−CD36+ phenotype (left, day 3). Quantification of CFU-E relative to shCTRL-transduced progenitors is presented (right, n = 9). (I) The differentiation of progenitors to an erythroid (GlyA) vs myeloid (CD11b) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative plots (left) and quantifications (right) are presented. Surface expression of GlyA and CD11b was monitored on shCTRL- and shSLC7A1-transduced progenitors at day 3 of differentiation (left). Quantification of erythroid (n = 14) and myeloid (n = 12) differentiation is shown in independent experiments (right). (J) CD34+CD38− progenitors transduced with shCTRL and shCAT1 vectors were evaluated for CFU potential using the StemMACS HSC-CFU Assay Kit and the numbers of burst-forming unit erythroid (BFU-E) and CFU–granulocyte-macrophage (GM) colonies generated at day 14 are presented for 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. IgG, immunoglobulin G; ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/20/10.1182_blood.2022017584/2/m_blood_bld-2022-017584-gr1.jpeg?Expires=1765954225&Signature=B7ro2VoVwe8zbE4EoYGwkuTHqcdXN0-V2y4I2hgz3TCTSTpT20KbRi0JJeWjLohqQpCQFXbk54obWUtE~p4Vaq5r1N2UoNqAym70XIPyy8hHCvF0Uck9d-2xWy3PUCAZvhRA8cDqQvTiVTLlxMSAyu9h~wOgZyBW5mTEjr10DibeIZKmQAXF8DJ4DtD-bA40U878tbMvSW80RbhsGjbrbPxyH~TImYg33lzCxjLqt8dCeNXYiUikiOgVh0SYmtkM2aCRRNziU1gwxlsXch0cUC~21ZDWAXN2URxgRthzukWKc6Et1lRLW6N~bhFkNJtlszrQu6o3PC6YuemFUqnPug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Erythroid differentiation is dependent on arginine-derived polyamine biosynthesis but not transport. (A) Schematic representation of polyamine biosynthesis from arginine with key enzymes indicated in red (ornithine carboxylase [ODC], spermidine synthase [SRM], spermine synthase [SMS], spermidine/spermine-N1-acetyl transferase [SAT1]). Steps that are inhibited by α-difluoromethylornithine (DMFO), trans-4-methyl cyclohexylamine (MCHA), and N-(3-amino-propyl)cyclohexylamine (APCHA) are presented. N1, N11-diethylnorspermine (DENS), an activator of polyamine catabolism, is also indicated. (B) The impact of DFMO (1 mM) on rEPO-induced differentiation of CD34+ progenitors was monitored by evaluating CD11b/GlyA profiles at days 3 and 7. Representative dot plots (top) and quantification of relative levels at days 3 (n = 14 for GlyA, n = 12 for CD11b) and 7 (n = 11 for GlyA, n = 9 for CD11b) of differentiation are shown (bottom). (C) The impact of MCHA (100 μM) was evaluated as a function of CD11b/GlyA profiles (top) at day 3 and quantifications are shown (bottom, n = 7). (D) The impact of DENS (10 μM) on EPO-induced differentiation was evaluated at day 3 and representative plots (top) and quantifications (n = 7 for GlyA and n = 5 for CD11b) are shown (bottom). (E) APCHA (100 μM) was added to rEPO-induced progenitors and representative profiles (top) and quantifications (bottom, n = 7) are shown. (F) DFMO-treated progenitors were differentiated with rEPO in the presence or absence of putrescine (Put; 100 μM) or spermidine (Spd; 100 μM), and GlyA was evaluated at day 7 (top, green histograms). Quantification from 7 independent experiments is presented (bottom). (G) Erythroid differentiation was induced in the presence or absence of DFMO, spermidine, and AMXT-1501 (2.5 μM), an inhibitor of polyamine transport. Quantification of GlyA expression relative to control conditions is presented (bottom, n = 3). ∗P < .05; ∗∗P < .001; ∗∗∗P < .001; ∗∗∗∗P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/20/10.1182_blood.2022017584/2/m_blood_bld-2022-017584-gr3.jpeg?Expires=1765954225&Signature=PrYgPPPQA4GO1~ITmgXDQQ-J1GKLOuAMyrlYZeh36yISZ8QyA2YvYQR8uOba~eeeYopT3HuVw0Sb-idMm0CnOiwALHqjs4IXMKA9LYtEQg7-HJ7lRVIi5rBM0fRUu3mT7lFjPEtitRiwZu-~y2PKE8dYZV5qHt7vu4JzxH04TDYOoWq8QvW2iry4RBN4xQD26yE7y1ANPZejcJWQ77oQq2DF-FPs8gVoTEyFlcLBQJaYdr4n7J3lVvNdJc62wFVyO1a8Rbc8yRokNRvpOpkTtHKATMjasffWsT7hYMF2ycJexklbjbikdnP4PDDBNQpCJcd1VPqKbe0UqDbombijoQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
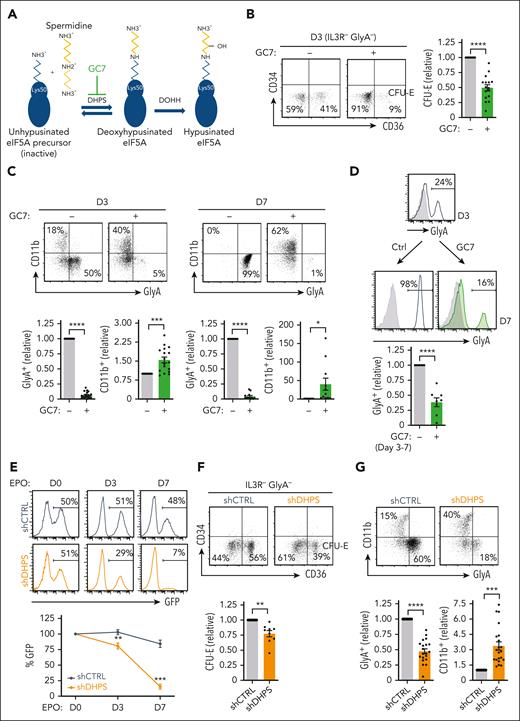
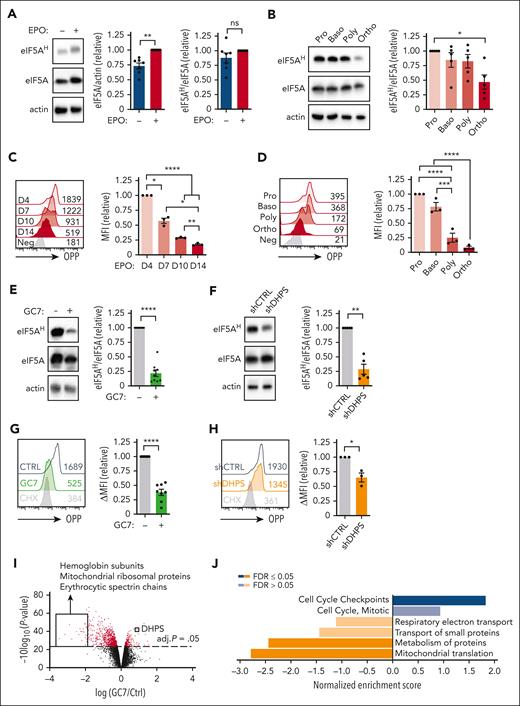
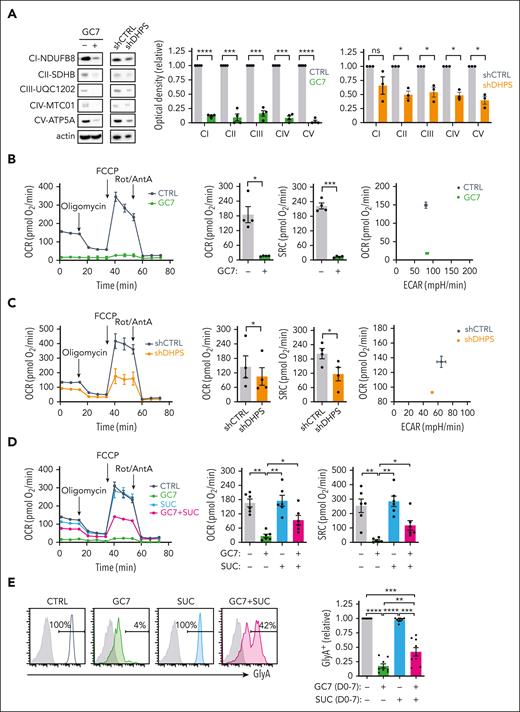
![Haploinsufficiency in RP genes is coupled to an attenuated hypusination in erythroid progenitors. (A) CD34+ BM progenitors from a healthy control and a patient with del(5q)-MDS were expanded ex vivo. Immunoblots showing hypusinated and total eIF5A levels at days 0 and 3 of EPO-induced differentiation are presented (left). Hypusination in CD34+ BM progenitors from a second patient with del(5q)-MDS was compared with progenitors from a patient with refractory anemia, after a 3-day differentiation in the absence (−) or presence (+) of EPO and blots are shown (middle). Quantification of the ratios of eIF5A/actin and eIF5AH/eIF5A for progenitors from 3 patients with del(5q)-MDS are presented (n = 3). (B) CD34+ BM progenitors from a RPL11-haploinsufficient patient with DBA [c.27_28 delGA (p.N10FS)] were evaluated for hypusinated and total eIF5A levels at isolation (day 0) and then expanded for 4 days followed by 3 days of EPO-induced differentiation (left). The change in eIF5AH/eIF5A after differentiation is shown. Peripheral blood (PB) CD34+ progenitors from a second RPL11-haploinsufficient patient (c.164dup; p.Tyr55Ter) were evaluated after ex vivo differentiation with a 4-day expansion followed by a 3 day EPO stimulation. eIF5AH immunoblots are shown (middle). Quantifications of the ratios of eIF5A/actin and eIF5AH/eIF5A for the 2 patients with DBA and 2 controls at day 3 of EPO-induced differentiation are presented (right). (C) The differentiation of shCTRL- and shRPL11-transduced progenitors to a vs n erythroid (GlyA+) vs myeloid (CD11b+) fate was evaluated by flow cytometry at day 3 of rEPO-induced differentiation. Representative CD11b/GlyA dot plots (left) and quantifications are presented (right, n = 10). (D) Hypusination was evaluated in human progenitors transduced with shCTRL and shRPL11 vectors at day 3 of differentiation. Representative immunoblots of eIF5AH and total eIF5A levels in shCTRL- and shRPL11-transduced progenitors (left) and quantifications ± SEM of eIF5AH/eIF5A and eIF5A/actin are presented (right, n = 6). Levels in control cells are set at “1.” (E) OCRs of shCTRL- and shRPL11-transduced progenitors were evaluated at day 1 of differentiation and representative data (left, n = 3-6 technical replicates) and mean basal OCR levels ± SEM are presented (right, n = 3 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/20/10.1182_blood.2022017584/2/m_blood_bld-2022-017584-gr7.jpeg?Expires=1765954225&Signature=TSoh4MkpeJwkUn8nLnUA9H6UTWpC4D87qcc7b4~~GMuwMB6s9fYbPbiTktkA5PAYpXdraWNraxa3YU1ZZci8mDJYyqbFsqZ1ZKzkOZyXr8Vgzo-012fqKwZSvB3VSeHK-tRODMppYbgeEUKH0gXBtL5FlBBO2a6oSmcamiUtfRKbFsJcwehhmj4-FVCoyRCQasWvgekMx2iymSgItq0GW7hhceZsyISyvVWeVALPYCmGRmaZeBl0Tb6uFXCrC-Vp6KEgOTg0ne~UQgOkkJq2eu88J9QSNqyeumelLDLvFHs28H-esWBXw8T-xwyagGHmF2j1YBclFfds5lIhawN~yA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal