Key Points
Molecular mechanism underlying autoantibody-mediated inhibition of ADAMTS13 activity in iTTP is not fully understood.
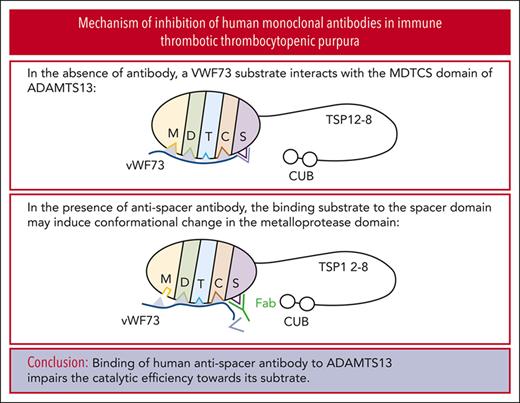
We found that binding of human antispacer antibody to ADAMTS13 results in conformational changes in its catalytic domain.
Abstract
Antibody binding to a plasma metalloprotease, a disintegrin and metalloproteinase with thrombospondin type 1 repeats 13 (ADAMTS13), is necessary for the development of immune thrombotic thrombocytopenic purpura (iTTP). Inhibition of ADAMTS13-mediated von Willebrand factor (VWF) cleavage by such antibodies clearly plays a role in the pathophysiology of the disease, although the mechanisms by which they inhibit ADAMTS13 enzymatic function are not fully understood. At least some immunoglobulin G–type antibodies appear to affect the conformational accessibility of ADAMTS13 domains involved in both substrate recognition and inhibitory antibody binding. We used single-chain fragments of the variable region previously identified via phage display from patients with iTTP to explore the mechanisms of action of inhibitory human monoclonal antibodies. Using recombinant full-length ADAMTS13, truncated ADAMTS13 variants, and native ADAMTS13 in normal human plasma, we found that, regardless of the conditions tested, all 3 inhibitory monoclonal antibodies tested affected enzyme turnover rate much more than substrate recognition of VWF. Hydrogen-to-deuterium exchange plus mass spectrometry experiments with each of these inhibitory antibodies demonstrated that residues in the active site of the catalytic domain of ADAMTS13 are differentially exposed to solvent in the presence and absence of monoclonal antibody binding. These results support the hypothesis that inhibition of ADAMTS13 in iTTP may not necessarily occur because the antibodies directly prevent VWF binding, but instead because of allosteric effects that impair VWF cleavage, likely by affecting the conformation of the catalytic center in the protease domain of ADAMTS13. Our findings provide novel insight into the mechanism of autoantibody-mediated inhibition of ADAMTS13 and pathogenesis of iTTP.
Introduction
Immune thrombotic thrombocytopenic purpura (iTTP) is caused by autoantibodies against the plasma metalloprotease, a disintegrin and metalloproteinase with thrombospondin type 1 repeats 13 (ADAMTS13).1-3 ADAMTS13 cleaves von Willebrand factor (VWF), preventing the accumulation of ultralarge VWF multimers that may occlude blood vessels and lead to organ damage.4 Severe deficiency of plasma ADAMTS13 activity results in iTTP, which presents with severe thrombocytopenia, microangiopathic hemolytic anemia, and organ dysfunction.5
Most patients with iTTP develop antibodies against the spacer domain of ADAMTS13, which functions as an exosite for binding of the VWF substrate.6-12 Although increased ADAMTS13 turnover caused by antibodies partially accounts for the pathophysiology of the disease, inhibitory antibodies, most of which target the spacer domain, clearly play a role in the disease process.7-9,13,14 Plasma or immunoglobulin G (IgG) from patients with iTTP can inhibit ADAMTS13 from normal human plasma (NHP) in most cases, demonstrating that functional inhibition is central to the pathophysiology of iTTP.15-19 As such, a better understanding of the disease will require more knowledge about the mechanism of IgG inhibitory antibodies.
It has widely been assumed that the inhibitory antispacer antibodies likely preclude the binding of VWF to ADAMTS13, although this hypothesis has not been tested extensively. Recent data suggest that anti-ADAMTS13 antibodies likely confer allosteric effects that can be observed distal to the putative binding epitopes. For example, a cryptic epitope on the spacer domain that is normally only accessible in the absence of the C-terminal domains of ADAMTS13 can be exposed by a mouse antihuman monoclonal antibody that binds to the C-terminal domains.6 Recent work by our group and others, also shows that antibodies that bind the distal C-terminal domains can stimulate the rate of ADAMTS13-mediated VWF cleavage, with some data suggesting that this is largely because of an increase in the catalytic rate, or turnover number, of the enzyme rather than the enhancement of substrate binding.20,21 However, the underlying assumptions allowing for such Michaelis-Menten–based kinetic analyses are rarely tested.22,23 Despite these limitations, available data qualitatively demonstrate that allosteric effects on the metalloprotease domain may be communicated via the entire length of the protein.6,20 Identifying the mechanism(s) by which inhibitors prevent enzymatic action will help determine whether it is feasible to develop technologies that prevent or rescue inhibition of ADAMTS13 in iTTP. Similarly, elucidation of the manner(s) in which antibodies prevent ADAMTS13-mediated VWF cleavage may also have a diagnostic value. A better understanding of the interplay of inhibitors in this polyclonal disease, such as iTTP certainly will lead to the development of better predictive and prognostic tools.
Here, we performed titrations of ADAMTS13, both in recombinant and native forms, with a fluorogenic surrogate substrate in the presence of a panel of human monoclonal antibodies previously identified via phage display.7,24,25 Surprisingly, we found that all 3 inhibitory antibodies that we tested likely affect the catalytic rate of VWF cleavage by ADAMTS13. These results were supported by hydrogen-to-deuterium exchange mass spectrometry (HX-MS) experiments, with all 3 inhibitors showing differential solvent exposure in residues at, or near, the active site of the metalloprotease domain despite the antibodies bound to the distant spacer domain.24 Our study is the first to test the inhibitory mechanism of anti spacer domain antibodies, which brings new insights into the pathophysiology of iTTP.
Methods
Purification of scFvs
Constructs, comprising a pET28 expression vector containing the coding sequences of the single-chain fragment of the variable region (scFv)4-20, scFv3-1, and scFv4-16, were prepared commercially (Twist Bioscience, South San Francisco, CA). The constructs were transformed into BL21 cells for expression as previously described.24 Concentration was determined by absorbance at 280 nM (NanoDrop, ThermoFisher, Waltham, MA). Purity was determined to be >95% by both Coomassie blue and Ponceau staining methods for each scFv used for functional assays.26,27
Production of recombinant ADAMTS13 and its variants
Recombinant full-length ADAMTS13 and truncated variants (MDTCS and MDTCS-T4) were prepared as previously described.28 MDTCS consists of the first 5 N-terminal domains of ADAMTS13 including the metalloprotease domain (M), disintegrin domain (D), the first thrombospondin domain (T), cysteine-rich domain (C), and spacer domain (S). MT4 is the MDTCS variant plus 3 additional thrombospondin repeats (T2-T4). Final products were concentrated with a spin concentrator with a cutoff chosen based on the size of the desired protein, and buffer exchanged to 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.45, 150 mM NaCl. Concentration was determined by absorbance of 280 nm using a NanoDrop.
ADAMTS13 functional assays
Proteolytic activity of plasma or recombinant ADAMTS13 and its truncated variants was tested by fluorescence of emission reflecting the cleavage of a 5′-maleimide fluorescein-labeled recombinant VWF73 fragment, known as fluorescence resonance energy transfer substrate (FRETS)-VWF73.21,29,30 Activity was determined by measuring the maximum change in fluorescence per unit time within the first 10 minutes of the reaction, with velocities reported as the change in fluorescence units per second (ΔFU/s). Depending on the experiment, scFvs were incubated at different concentrations with 2.5 μL pooled NHP (Sigma Aldrich, St Louis, MO) containing wild-type ADAMTS13, or purified recombinant ADAMTS13 and truncated variants at a concentration ranging between 1.5 and 2 μg/mL, for 15 minutes at 37°C, before addition of FRETS-VWF73. The reaction system was in a buffer containing 5 mM Bis-Tris, pH 6.0, 25 mM CaCl2, 0.005% Tween 20, and 1 mM Pefabloc (Sigma Aldrich) with a pH of either at 6.0 or at 6.31, in a 96-well opaque white Nunc plate (ThermoFisher), and measured on a SpectraMax Gemini XPS plate reader (Molecular Devices, San Jose, CA) at a temperature of either 25 or 37°C after the addition of substrate to the wells.
For analysis of the reversibility of inhibition by the scFvs, the cleavage activity (ΔFU/s) of NHP or a full-length recombinant ADAMTS13 was measured in the absence of scFv4-20 with a FRETS-VWF73 concentration of 1.0 μM. Recombinant ADAMTS13 and variant concentrations used were the same as described above. Enzyme activity in the presence of scFv4-20 at a concentration at, or near, the 50% inhibitory concentration, as determined by separate titrations at the same substrate concentration, was then compared with uninhibited enzyme. Each time point represents incubation of enzyme with scFv4-20 at 37°C before addition of substrate. Activity was normalized to the velocity of uninhibited enzyme at each time point as well as to fully inhibited enzyme using ethylenediaminetetraacetic acid (EDTA) at a final concentration of 20 mM. For titration of ADAMTS13 with substrate, scFv3-1, scFv4-20, or scFv4-16 were incubated with NHP or recombinant ADAMTS13 or truncated variants before addition of FRETS-VWF73 over a range of final substrate concentrations as described in the text.
Enzyme kinetics analysis
After reaction velocities were determined using Microsoft Excel, the maximum enzyme velocity (Vmax) of ADAMTS13 activity and the substrate concentration at which enzyme velocity is at the half-maximal (K0.5) were calculated by hyperbolic fit (GraphPad Prism 9, San Diego, CA), as previously described.21 Data sets with the same enzyme and reaction conditions but different fixed concentrations of antibodies were fit to an equation derived from Michaelis-Menten kinetics (Equation 1, supplemental Figure 1, available on the Blood website). The analysis is based on Michaelis-Menten kinetics, because the assumption of pseudo–first order kinetics cannot be made based on previously available data as well as the enzymatic behavior of ADAMTS13 observed here.21,31 The parameter αapp, which accounts for the apparent effect of nonsubstrate ligand on K0.5, represents the relative effect of an inhibitor on substrate binding, as elaborated upon in the supplemental Materials.22,23,32 Deviation of αapp from unity (αapp = 1), particularly when deviation approaches an order of magnitude in either direction, is evidence of the effect of an inhibitor on substrate binding.
HX-MS
Hydrogen-to-deuterium exchange in the MDTCS fragment was measured by mass spectrometry following enzyme fragmentation and separation in the absence or presence of each of scFvs, as previously described.24 Briefly, for an antibody-bound MDTCS, scFv was immobilized on Affi-Gel15 resin (Bio-Rad, Hercules, CA) following the manufacturer’s protocol, packed into a steel (2 mm × 2 cm) column; the MDTCS was allowed to bind to the column; a D2O buffer was flushed into the column. Deuterium-labeled MDTCS was eluted from the column with an acid buffer (pH 2.5) and injected into an immobilized pepsin protease column. After exposure to a small C8 trap column and separation using an analytical C18 column, peptides were analyzed by mass spectrometry (LTQ Orbitrap XL, ThermoFisher). Comparative experiments were done with MDTCS in free solution, as previously described.24 Peptides were identified and analyzed for deuterium labeling by the HDExaminer program (Sierra Analytics, Modesto, CA) at peptide resolution; results were confirmed by analysis using the program ExMS2.33
Results
Antibody-mediated inhibition affects catalytic turnover
To explore how an inhibitory antibody would prevent ADAMTS13-mediated VWF cleavage, we determined the effects of a well-characterized inhibitory antibody fragment scFv4-20 on the recombinant versions of ADAMTS13 in a FRETS-based assay (Figure 1A). The binding site of scFv4-20 has been identified on the spacer domain and it potently inhibits both recombinant ADAMTS13 and plasma ADAMTS13.7,21,24 The potency and purity of scFv4-20 and other antibodies described in further sections were verified using a functional assay and sodium dodecyl sulfate–polyacrylamide gel (SDS-PAGE) with Coomassie blue staining, respectively (not shown).
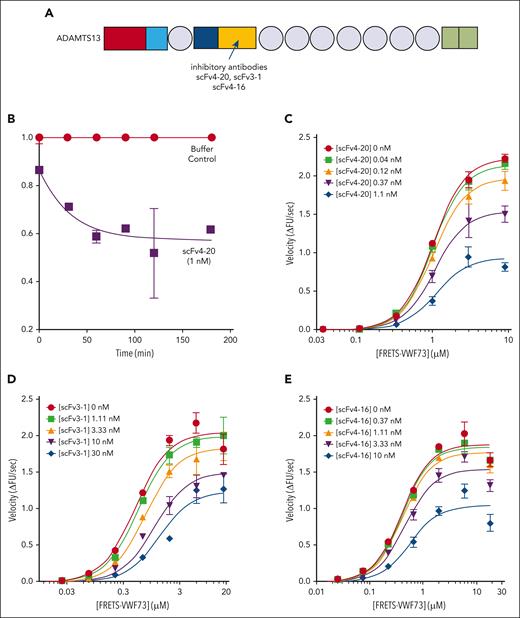
Kinetic inhibition of recombinant full-length ADAMTS13 and truncated variants by an antispacer antibody (scFv20). (A) Cartoon representation of the structure of ADAMTS13 and truncated variants. Each recombinant protein has a 6-His tag and a V5 tag at the C-terminus. Rectangles: red, metalloprotease domain; light blue, disintegrin domain; dark blue, cysteine-rich domain; yellow, spacer domain; and green, 2 complement C1r/C1s, Uegf, Bmp1 (CUB) domains. Circles: thrombospondin type-1 domains (T2-8). Spacer domain, the binding site of scFv4-20 to ADAMTS13, is labeled. Assays were done at 25°C and pH 6.0. (B) The relative inhibitory activity of scFv4-20 incubated with full-length ADAMTS13 for different lengths of time (0, 15, 30, and 60 minutes). Activity was normalized to velocity (ΔFU/s) in the absence of scFv4-20 (buffer control) to derive relative velocity. (C-E) Titration of recombinant full-length ADAMTS13, truncated ADAMTS13 MT4, and MDTCS, respectively, with FRETS-VWF73 in the presence of various concentrations of scFv4-20 (n ≥ 3). Data in the plots are shown as mean ± standard error of the mean (SEM).
Kinetic inhibition of recombinant full-length ADAMTS13 and truncated variants by an antispacer antibody (scFv20). (A) Cartoon representation of the structure of ADAMTS13 and truncated variants. Each recombinant protein has a 6-His tag and a V5 tag at the C-terminus. Rectangles: red, metalloprotease domain; light blue, disintegrin domain; dark blue, cysteine-rich domain; yellow, spacer domain; and green, 2 complement C1r/C1s, Uegf, Bmp1 (CUB) domains. Circles: thrombospondin type-1 domains (T2-8). Spacer domain, the binding site of scFv4-20 to ADAMTS13, is labeled. Assays were done at 25°C and pH 6.0. (B) The relative inhibitory activity of scFv4-20 incubated with full-length ADAMTS13 for different lengths of time (0, 15, 30, and 60 minutes). Activity was normalized to velocity (ΔFU/s) in the absence of scFv4-20 (buffer control) to derive relative velocity. (C-E) Titration of recombinant full-length ADAMTS13, truncated ADAMTS13 MT4, and MDTCS, respectively, with FRETS-VWF73 in the presence of various concentrations of scFv4-20 (n ≥ 3). Data in the plots are shown as mean ± standard error of the mean (SEM).
To verify the underlying assumption that scFv4-20 reversibly inhibits ADAMTS13 activity, we compared the enzyme activity of recombinant ADAMTS13 in the presence and absence of the inhibitor at different time points (Figure 1B). Because the concentration of the inhibitor in the assay is in excess of the concentration of the enzyme, if inhibition was irreversible, then the functional concentration of ADAMTS13 in the assay would decrease over time, as would the activity. Because inhibition plateaus at 15 minutes in these conditions, reversible inhibition can be assumed to occur based on the known range of kon of scFvs, supporting the use of a Michaelis-Menten–derived interpretation of results.34-36 Recombinant full-length ADAMTS13 (FL-A13) was titrated with an increasing concentration of FRETS-VWF73 substrate in the presence of an increasing concentration of inhibitor scFv4-20 (Figure 1C). The maximum enzyme velocity (Vmax) decreased by approximately threefold (1.05 ± 0.09 ΔFU/s vs 0.36 ± 0.02 ΔFU/s) whereas the substrate concentration at which velocity was half-maximal (K0.5) changed less than twofold (1.11 ± 0.05 μM vs 2.32 ± 0.18 μM) when comparing titrations in the absence of inhibitor and at the highest scFv4-20 concentration (Table 1). Similar results were observed when comparing the titrations in the absence of, and at the highest concentration of, scFv4-20 when using the truncated versions of recombinant ADAMTS13 with either the last 6 C-terminal domains deleted (MT4) (0.65 ± 0.02 ΔFU/s vs 0.11 ± 0.02 ΔFU/s; and 0.44 ± 0.02 μM vs 0.92 ± 0.35 μM) or all domains distal to the spacer domain deleted (MDTCS) (1.42 ± 0.12 ΔFU/s vs 0.26 ± 0.01 ΔFU/s; and 1.01 ± 0.08 μM vs 2.37 ± 0.26 μM) (Figure 1D-E; Table 1). These results suggested that altering the rate of catalytic turnover (kcat) may play an important role in the mechanism of inhibition of scFv4-20, and that the apparent allosteric effect does not depend on the domains distal to the spacer domain.
Kinetic parameters of scFv4-20–mediated inhibition of recombinant ADAMTS13 and variants
| Constructs . | scFv4-20 (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global fit) . |
|---|---|---|---|---|
| FL-AT13 | 0 | 1.05 ± 0.09 | 1.11 ± 0.05 | 1.82 ± 0.06 |
| 0.37 | 1.14 ± 0.08 | 1.45 ± 0.10 | ||
| 1.1 | 0.98 ± 0.09 | 1.22 ± 0.03 | ||
| 3.3 | 0.88 ± 0.08 | 1.71 ± 0.13 | ||
| 10 | 0.36 ± 0.02 | 2.32 ± 0.18 | ||
| MT4 | 0 | 0.65 ± 0.02 | 0.44 ± 0.02 | 1.27 ± 0.07 |
| 0.12 | 0.67 ± 0.07 | 0.56 ± 0.10 | ||
| 0.37 | 0.59 ± 0.04 | 0.50 ± 0.02 | ||
| 1.1 | 0.34 ± 0.05 | 0.50 ± 0.08 | ||
| 3.3 | 0.11 ± 0.02 | 0.92 ± 0.35 | ||
| MDTCS | 0 | 1.42 ± 0.12 | 1.01 ± 0.08 | 1.81 ± 0.39 |
| 1.1 | 1.55 ± 0.14 | 1.39 ± 0.14 | ||
| 3.3 | 1.27 ± 0.14 | 1.26 ± 0.04 | ||
| 10 | 0.89 ±0.09 | 1.77 ± 0.35 | ||
| 30 | 0.26 ± 0.01 | 2.37 ± 0.26 |
| Constructs . | scFv4-20 (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global fit) . |
|---|---|---|---|---|
| FL-AT13 | 0 | 1.05 ± 0.09 | 1.11 ± 0.05 | 1.82 ± 0.06 |
| 0.37 | 1.14 ± 0.08 | 1.45 ± 0.10 | ||
| 1.1 | 0.98 ± 0.09 | 1.22 ± 0.03 | ||
| 3.3 | 0.88 ± 0.08 | 1.71 ± 0.13 | ||
| 10 | 0.36 ± 0.02 | 2.32 ± 0.18 | ||
| MT4 | 0 | 0.65 ± 0.02 | 0.44 ± 0.02 | 1.27 ± 0.07 |
| 0.12 | 0.67 ± 0.07 | 0.56 ± 0.10 | ||
| 0.37 | 0.59 ± 0.04 | 0.50 ± 0.02 | ||
| 1.1 | 0.34 ± 0.05 | 0.50 ± 0.08 | ||
| 3.3 | 0.11 ± 0.02 | 0.92 ± 0.35 | ||
| MDTCS | 0 | 1.42 ± 0.12 | 1.01 ± 0.08 | 1.81 ± 0.39 |
| 1.1 | 1.55 ± 0.14 | 1.39 ± 0.14 | ||
| 3.3 | 1.27 ± 0.14 | 1.26 ± 0.04 | ||
| 10 | 0.89 ±0.09 | 1.77 ± 0.35 | ||
| 30 | 0.26 ± 0.01 | 2.37 ± 0.26 |
All recombinant proteins expressed in human embryonic kidney (HEK) 293 cells. ScFv4-20 is a single-chain fragment of a variable loop that binds to the spacer domain of ADAMTS13. Reaction conditions: temperature 25°C, pH 6.0. All data are presented as mean ± SEM.
Here, FL-AT13, full-length recombinant ADAMTS13; FU, fluorescence units; MDTCS, a truncated recombinant ADAMTS13 consisting of the first 5 N-terminal domains; MT4, a truncated recombinant ADAMTS13 including MDTCS plus the first 4 TSP1-like repeats.
Next, we endeavored to determine the extent to which scFv4-20 may affect substrate concentration–related effects on ADAMTS13 activity. When the curves were fit to a single Michaelis-Menten–derived equation (see “Methods”), the relative effect of the inhibitor on K0.5 was quantified (αapp), with values close to 1 suggesting little to no effect on K0.5.22,23,32 The presence of clear plateaus of activity both at high and low substrate concentrations further supports the validity of the approach used to derive the αapp term (Figure 1). In each case, both here and as described in further sections for all additional experiments, the value of αapp was consistently within half an order of magnitude of 1 (1.82 ± 0.06 for FL-A13, 1.27 ± 0.07 for MT4, and 1.81 ± 0.39 for MDTCS) (Table 1).
Inhibitory antibodies decrease plasma ADAMTS13 catalytic rate
To determine how inhibitory antibodies affect human ADAMTS13 in plasma, we observed the cleavage rates of FRETS-VWF73 in the presence of scFv4-20 as well as 2 additional previously described inhibitory antibodies from the same phage display library that also target the spacer domain (eg, scFv3-1 and scFv4-16) (Figure 2A).7,24 To ensure that the inhibition was reversible in these conditions, we observed inhibition at different time points in the presence and absence of scFv4-20; the inhibition plateaued at 30 minutes (Figure 2B).
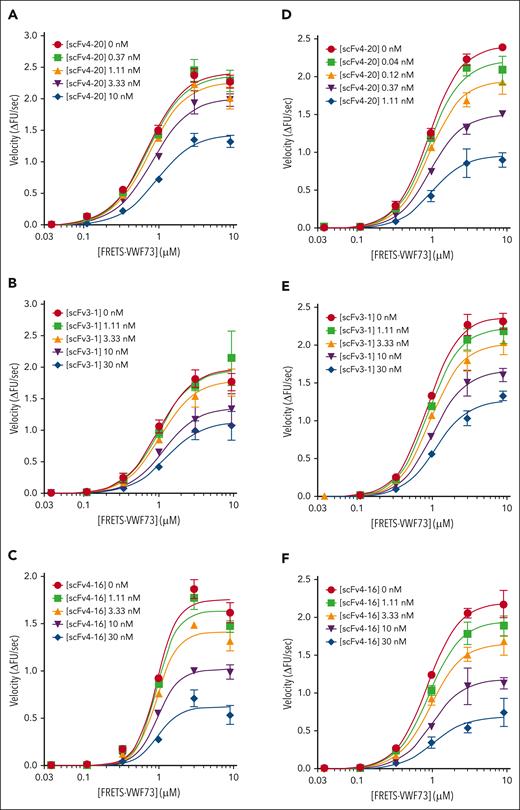
Kinetic inhibition of plasma ADAMTS13 by various antispacer antibodies (scFv20, scFv3-1, and scFv4-16). (A) Cartoon representation of the structure of ADAMTS13, as described in Figure 1. Spacer domain, the binding site of scFv4-20, scFv3-1, and scFv4-16 to ADAMTS13, is indicated by an arrow. Assays were performed at 25°C and pH 6.0. (B) Relative inhibitory activity variation of scFv4-20 incubated with NHP for different lengths of time (0, 30, 60, 90, 120, and 180 minutes). Activity was normalized to velocity (ΔFU/s) in the absence of scFv4-20 (buffer control) to derive relative velocity. (C-E) The titrations of ADAMTS13 in NHP with FRETS-VWF73 in the presence of increasing concentrations of scFv4-20, scFv3-1, and scFv4-16, respectively (n ≥ 3 for all titrations). The data in the plots are shown as mean ± SEM.
Kinetic inhibition of plasma ADAMTS13 by various antispacer antibodies (scFv20, scFv3-1, and scFv4-16). (A) Cartoon representation of the structure of ADAMTS13, as described in Figure 1. Spacer domain, the binding site of scFv4-20, scFv3-1, and scFv4-16 to ADAMTS13, is indicated by an arrow. Assays were performed at 25°C and pH 6.0. (B) Relative inhibitory activity variation of scFv4-20 incubated with NHP for different lengths of time (0, 30, 60, 90, 120, and 180 minutes). Activity was normalized to velocity (ΔFU/s) in the absence of scFv4-20 (buffer control) to derive relative velocity. (C-E) The titrations of ADAMTS13 in NHP with FRETS-VWF73 in the presence of increasing concentrations of scFv4-20, scFv3-1, and scFv4-16, respectively (n ≥ 3 for all titrations). The data in the plots are shown as mean ± SEM.
NHP containing a wild-type ADAMTS13 was titrated with an increasing concentration of FRETS-VWF73 in the presence of fixed concentrations of scFv4-20 (Figure 2C). The Vmax decreased with increasing concentrations of inhibitor (2.79 ± 0.07 ΔFU/s vs 1.10 ± 0.08 ΔFU/s) comparing absent scFv4-20 and highest concentration used, whereas the K0.5 was nearly identical (1.71 ± 0.05 μM vs 1.60 ± 0.12 μM); the parameter αapp was determined to be 1.20 ± 0.10 (Table 2). Titration of NHP with FRETS-VWF73 in the presence of fixed concentrations of scFv3-1 also showed a decrease in Vmax comparing absent inhibitor and highest concentration used (2.19 ± 0.12 ΔFU/s vs 1.56 ± 0.28 ΔFU/s); K0.5, however, did increase by approximately fourfold (0.58 ± 0.07 μM vs 2.46 ± 0.52 μM) (Figure 2D). Global fit of the scFv3-1 data revealed an αapp value of 2.78 ± 0.26. Titration of NHP with FRETS-VWF73 in the presence of fixed concentrations of scFv4-16 showed a decrease in Vmax (2.01 ± 0.06 ΔFU/s vs 1.11 ± 0.08 ΔFU/s) and very little change in K0.5 (0.45 ± 0.02 μM vs 0.63 ± 0.08 μM) when comparing data without scFv4-16 and at the highest concentration tested, with an αapp value of 1.61 ± 0.03 (Figure 2E). To this point, experiments were done under standard assay conditions, as previously described (pH, 6.0; 25°C).21,24
Kinetic inhibition of native ADAMTS13 in plasma with various antispacer domain inhibitors
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.79 ± 0.07 | 1.71 ± 0.05 | 1.20 ± 0.10 |
| 0.04 | 2.72 ± 0.08 | 1.69 ± 0.11 | ||
| 0.12 | 2.45 ± 0.13 | 1.75 ± 0.19 | ||
| 0.37 | 1.97 ± 0.14 | 1.94 ± 0.33 | ||
| 1.1 | 1.10 ± 0.09 | 1.60 ± 0.12 | ||
| scFv3-1 | 0 | 2.19 ± 0.12 | 0.58 ± 0.07 | 2.78 ± 0.26 |
| 1.1 | 2.18 ± 0.11 | 0.77 ± 0.13 | ||
| 3.3 | 1.97 ± 0.04 | 0.87 ± 0.08 | ||
| 10 | 1.56 ± 0.10 | 1.12 ± 0.21 | ||
| 30 | 1.56 ± 0.28 | 2.46 ± 0.52 | ||
| scFv4-16 | 0 | 2.01 ± 0.06 | 0.45 ± 0.02 | 1.61 ± 0.13 |
| 0.37 | 1.95 ± 0.03 | 0.46 ± 0.03 | ||
| 1.1 | 1.91 ± 0.06 | 0.49 ± 0.04 | ||
| 3.3 | 1.64 ±0.03 | 0.53 ± 0.03 | ||
| 10 | 1.11 ± 0.08 | 0.63 ± 0.08 |
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.79 ± 0.07 | 1.71 ± 0.05 | 1.20 ± 0.10 |
| 0.04 | 2.72 ± 0.08 | 1.69 ± 0.11 | ||
| 0.12 | 2.45 ± 0.13 | 1.75 ± 0.19 | ||
| 0.37 | 1.97 ± 0.14 | 1.94 ± 0.33 | ||
| 1.1 | 1.10 ± 0.09 | 1.60 ± 0.12 | ||
| scFv3-1 | 0 | 2.19 ± 0.12 | 0.58 ± 0.07 | 2.78 ± 0.26 |
| 1.1 | 2.18 ± 0.11 | 0.77 ± 0.13 | ||
| 3.3 | 1.97 ± 0.04 | 0.87 ± 0.08 | ||
| 10 | 1.56 ± 0.10 | 1.12 ± 0.21 | ||
| 30 | 1.56 ± 0.28 | 2.46 ± 0.52 | ||
| scFv4-16 | 0 | 2.01 ± 0.06 | 0.45 ± 0.02 | 1.61 ± 0.13 |
| 0.37 | 1.95 ± 0.03 | 0.46 ± 0.03 | ||
| 1.1 | 1.91 ± 0.06 | 0.49 ± 0.04 | ||
| 3.3 | 1.64 ±0.03 | 0.53 ± 0.03 | ||
| 10 | 1.11 ± 0.08 | 0.63 ± 0.08 |
Each inhibitor binds to the spacer domain of ADAMTS13. Reaction conditions: temperature 25°C, pH 6.0. All data are presented as mean ± SEM.
To determine whether temperature- or pH-related effects affect the mechanism of inhibition, we repeated the titrations as described in Figure 2 at a higher temperature and pH, respectively, which more closely reflects physiologic conditions (Figure 3). In conditions of pH 6.0 and 37°C, the Vmax decreased for each inhibitor tested, comparing absent inhibitor and the highest fixed concentration (2.73 ± 0.12 ΔFU/s vs 1.64 ± 0.12 ΔFU/s for scFv4-20; 2.21 ± 0.21 ΔFU/s vs 1.50 ± 0.33 ΔFU/s for scFv3-1; and 2.11 ± 0.15 ΔFU/s vs 1.76 ± 0.11 ΔFU/s for scFv4-16); the K0.5 increased by less than twofold for each inhibitor (0.92 ± 0.08 μM vs 1.28 ± 0.08 μM for scFv4-20; 1.29 ± 0.29 μM vs 2.39 ± 0.81 μM for scFv3-1; and 1.28 ± 0.14 μM vs 1.44 ± 0.26 μM for scFv4-16); and the value of αapp was less than 2 for each inhibitor (1.71 ± 0.07 for scFv4-20, 1.17 ± 0.15 for scFv3-1, and 1.06 ± 0.06 for scFv4-16) (Table 3). In conditions of pH 6.31 and 25°C, the Vmax decreased for each inhibitor tested, comparing absent inhibitor and the highest fixed concentration (2.96 ± 0.05 ΔFU/s vs 1.14 ± 0.18 ΔFU/s for scFv4-20; 2.88 ± 0.15 ΔFU/s vs 1.72 ± 0.07 ΔFU/s for scFv3-1; and 2.67 ± 0.20 ΔFU/s vs 1.00 ± 0.29 ΔFU/s for scFv4-16); the K0.5 increased by less than twofold for each inhibitor (1.48 ± 0.03 μM vs 1.61 ± 0.20 μM for scFv4-20; 1.35 ± 0.08 μM vs 2.36 ± 0.32 μM for scFv3-1; and 1.36 ± 0.11 μM vs 2.45 ± 1.04 μM for scFv4-16); and the value of αapp was less than 2 for each inhibitor (1.09 ± 0.17 for scFv4-20, 1.42 ± 0.22 for scFv3-1, and 1.27 ± 0.19 for scFv4-16) (Table 4). Use of pH values even closer to physiologic pH was not possible for these experiments because ADAMTS13 activity in vitro decreases significantly as conditions approach neutrality and does not produce sufficient signal to resolve the effects of these inhibitors on enzyme activity.21,37,38
Kinetic inhibition of plasma ADAMTS13 by three antispacer antibodies at different temperatures and pHs. Titration of plasma ADAMTS13 activity with FRETS-VWF73 in the presence of various concentrations of scFv4-20, scFv3-1, and scFv4-16 under different conditions: (1) temperature of 37°C and pH of 6.0 for scFv4-20 (A), scFv3-1 (B), and scFv4-16 (C); (2) temperature of 25°C and pH of 6.31 for scFv4-20 (D), scFv3-1 (E), and scFv4-16 (F) (n ≥ 3 for all titrations). The data in the plots are shown as mean ± SEM.
Kinetic inhibition of plasma ADAMTS13 by three antispacer antibodies at different temperatures and pHs. Titration of plasma ADAMTS13 activity with FRETS-VWF73 in the presence of various concentrations of scFv4-20, scFv3-1, and scFv4-16 under different conditions: (1) temperature of 37°C and pH of 6.0 for scFv4-20 (A), scFv3-1 (B), and scFv4-16 (C); (2) temperature of 25°C and pH of 6.31 for scFv4-20 (D), scFv3-1 (E), and scFv4-16 (F) (n ≥ 3 for all titrations). The data in the plots are shown as mean ± SEM.
Kinetic inhibition of plasma ADAMTS13 at 37°C and pH of 6.0
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.73 ± 0.12 | 0.92 ± 0.08 | 1.71 ± 0.07 |
| 0.37 | 2.80 ± 0.12 | 1.00 ± 0.06 | ||
| 1.1 | 2.49 ± 0.12 | 0.93 ± 0.11 | ||
| 3.3 | 2.41 ± 0.10 | 1.23 ± 0.11 | ||
| 10 | 1.64 ± 0.12 | 1.28 ± 0.08 | ||
| scFv3-1 | 0 | 2.21 ± 0.21 | 1.29 ± 0.29 | 1.17 ± 0.15 |
| 1.1 | 2.94 ± 0.85 | 2.49 ± 1.26 | ||
| 3.3 | 2.22 ± 0.38 | 1.74 ± 0.50 | ||
| 10 | 1.80 ± 0.40 | 1.65 ± 0.55 | ||
| 30 | 1.50 ± 0.33 | 2.39 ± 0.81 | ||
| scFv4-16 | 0 | 2.11 ± 0.15 | 1.28 ± 0.14 | 1.06 ± 0.06 |
| 1.1 | 1.93 ± 0.08 | 1.21 ± 0.10 | ||
| 3.3 | 1.70 ± 0.10 | 1.28 ± 0.10 | ||
| 10 | 1.26 ± 0.09 | 1.47 ± 0.25 | ||
| 30 | 1.76 ± 0.11 | 1.44 ± 0.26 |
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.73 ± 0.12 | 0.92 ± 0.08 | 1.71 ± 0.07 |
| 0.37 | 2.80 ± 0.12 | 1.00 ± 0.06 | ||
| 1.1 | 2.49 ± 0.12 | 0.93 ± 0.11 | ||
| 3.3 | 2.41 ± 0.10 | 1.23 ± 0.11 | ||
| 10 | 1.64 ± 0.12 | 1.28 ± 0.08 | ||
| scFv3-1 | 0 | 2.21 ± 0.21 | 1.29 ± 0.29 | 1.17 ± 0.15 |
| 1.1 | 2.94 ± 0.85 | 2.49 ± 1.26 | ||
| 3.3 | 2.22 ± 0.38 | 1.74 ± 0.50 | ||
| 10 | 1.80 ± 0.40 | 1.65 ± 0.55 | ||
| 30 | 1.50 ± 0.33 | 2.39 ± 0.81 | ||
| scFv4-16 | 0 | 2.11 ± 0.15 | 1.28 ± 0.14 | 1.06 ± 0.06 |
| 1.1 | 1.93 ± 0.08 | 1.21 ± 0.10 | ||
| 3.3 | 1.70 ± 0.10 | 1.28 ± 0.10 | ||
| 10 | 1.26 ± 0.09 | 1.47 ± 0.25 | ||
| 30 | 1.76 ± 0.11 | 1.44 ± 0.26 |
Each inhibitor binds to the spacer domain of ADAMTS13. All data are presented as mean ± SEM.
Kinetic inhibition of plasma ADAMTS13 at 25°C and pH of 6.31
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.96 ± 0.05 | 1.48 ± 0.03 | 1.09 ± 0.17 |
| 0.04 | 2.83 ± 0.15 | 1.44 ± 0.06 | ||
| 0.12 | 2.36 ± 0.22 | 1.46 ± 0.23 | ||
| 0.37 | 1.87 ± 0.05 | 1.66 ± 0.13 | ||
| 1.1 | 1.14 ± 0.18 | 1.61 ± 0.20 | ||
| scFv3-1 | 0 | 2.88 ± 0.15 | 1.35 ± 0.08 | 1.42 ± 0.22 |
| 1.1 | 2.71 ± 0.22 | 1.43 ± 0.14 | ||
| 3.3 | 2.51 ± 0.24 | 1.56 ± 0.19 | ||
| 10 | 2.03 ± 0.07 | 1.69 ± 0.21 | ||
| 30 | 1.72 ± 0.07 | 2.36 ± 0.32 | ||
| scFv4-16 | 0 | 2.67 ± 0.20 | 1.36 ± 0.11 | 1.27 ± 0.19 |
| 1.1 | 2.32 ± 0.18 | 1.38 ± 0.06 | ||
| 3.3 | 2.06 ± 0.21 | 1.45 ± 0.10 | ||
| 10 | 1.45 ± 0.13 | 1.69 ± 0.17 | ||
| 30 | 1.00 ± 0.29 | 2.45 ± 1.04 |
| Inhibitor . | Concentration (nM) . | Vmax (FU/s) . | K0.5 (μM) . | αapp (global-fit) . |
|---|---|---|---|---|
| scFv4-20 | 0 | 2.96 ± 0.05 | 1.48 ± 0.03 | 1.09 ± 0.17 |
| 0.04 | 2.83 ± 0.15 | 1.44 ± 0.06 | ||
| 0.12 | 2.36 ± 0.22 | 1.46 ± 0.23 | ||
| 0.37 | 1.87 ± 0.05 | 1.66 ± 0.13 | ||
| 1.1 | 1.14 ± 0.18 | 1.61 ± 0.20 | ||
| scFv3-1 | 0 | 2.88 ± 0.15 | 1.35 ± 0.08 | 1.42 ± 0.22 |
| 1.1 | 2.71 ± 0.22 | 1.43 ± 0.14 | ||
| 3.3 | 2.51 ± 0.24 | 1.56 ± 0.19 | ||
| 10 | 2.03 ± 0.07 | 1.69 ± 0.21 | ||
| 30 | 1.72 ± 0.07 | 2.36 ± 0.32 | ||
| scFv4-16 | 0 | 2.67 ± 0.20 | 1.36 ± 0.11 | 1.27 ± 0.19 |
| 1.1 | 2.32 ± 0.18 | 1.38 ± 0.06 | ||
| 3.3 | 2.06 ± 0.21 | 1.45 ± 0.10 | ||
| 10 | 1.45 ± 0.13 | 1.69 ± 0.17 | ||
| 30 | 1.00 ± 0.29 | 2.45 ± 1.04 |
Each inhibitor binds to the spacer domain of ADAMTS13. All data are presented as mean ± SEM.
Antispacer antibodies affect the conformation of the catalytic domain
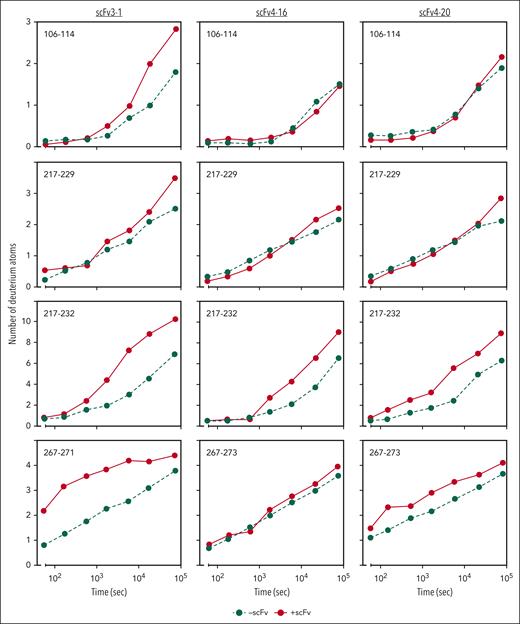
Previous work identified the ADAMTS13 spacer domain as the binding site of the inhibitors described herein. Immunoprecipitation experiments revealed that only variants of recombinant ADAMTS13 containing the spacer domain bind to scFv3-1, scFv4-16, and scFv4-20.7 Further HX-MS experiments identified the putative binding sites of the inhibitors on the spacer domain, with significant epitope overlap between all 3 scFvs tested.24 However, evidence of conformational change in other domains was not investigated and reported because a mechanistic interpretation of this data was not apparent to the investigators at the time, and multiple additional lines of evidence supported the authors’ claims that these inhibitors bind to the spacer domain. Functional data reported here suggest that antispacer domain antibodies may affect the metalloprotease domain in an allosteric manner. To explore this hypothesis, we carefully reviewed the previously collected HX-MS data of the MDTCS in the presence and absence of scFv3-1, scFv4-16, and scFv4-20. Altered levels of deuterium uptake in domains distal to a known binding epitope suggest allosteric activity. We found that deuterium uptake is higher in the metalloprotease domain in the presence of all 3 scFvs when compared with that in the absence of antibody binding. Of the 3 inhibitors, scFv3-1 demonstrated the most differences between the bound and unbound states within the metalloprotease domain, with differential deuterium uptake observed in peptide fragments including residues T106-L114, D217-A223, D217-E225, D217-S229, D217-F230, D217-L232, D217-E233, A261-L273, and R267-L273 (Figure 4; supplemental Figure 2). The inhibitor scFv4-20 showed differential uptake in peptide fragments containing residues D217-L232, D217-E233, A261-L273, and R267-L273 (Figure 4; supplemental Figure 2). For scFv4-16, differential uptake was seen in peptide fragments containing residues D217-L232 and D217-E233 (Figure 4; supplemental Figure 2). In addition, differential uptake in a peptide fragment containing residues I222-L232 was seen using scFv4-20, although this peptide fragment was not detected in experiments using scFv3-1 and scFv4-16 (data not shown). The observation of several overlapping peptides with similar uptake profiles, as well as accounting for the fact that deuterium label is lost for the first 1 or 2 residues after proteolysis,39 revealed that the residues most likely involved in conformational change were N107-L114, I222-A223, H228-L232, and W262-S272 for scFv3-1; H228-L232 and W262-S272 for scFv4-20; and H228-L232 for scFv4-16 (Figure 5). Of note, no overlapping peptides were available to compare the residues between T106-L114 in any of the experiments, and no peptides that included residues H234 or D235 were available for analysis using any of the scFvs. Together, the data demonstrate a long-range conformational change in the metalloprotease domain when an inhibitory antibody binds the spacer domain of ADAMTS13.
HX-MS reveals allosteric effects on the metalloprotease domain by antispacer antibodies. Deuterium incorporation of MDTCS fragments is measured over the time course of 21 hours with or without antibody bound. Incorporation in the absence of scFvs is depicted in green circles and dashed lines; incorporation of deuterium in the presence of scFvs is depicted in red circles and solid lines. The y-axis indicates the number of deuterium atoms incorporated over time on the x-axis.
HX-MS reveals allosteric effects on the metalloprotease domain by antispacer antibodies. Deuterium incorporation of MDTCS fragments is measured over the time course of 21 hours with or without antibody bound. Incorporation in the absence of scFvs is depicted in green circles and dashed lines; incorporation of deuterium in the presence of scFvs is depicted in red circles and solid lines. The y-axis indicates the number of deuterium atoms incorporated over time on the x-axis.
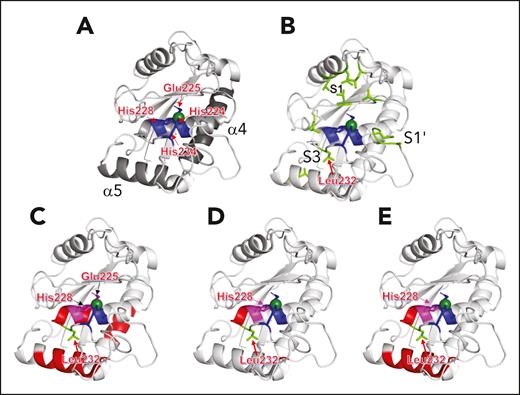
ADAMTS13 M domain residues exhibit a conformational change in the presence of antispacer domain inhibitors. (A) Depiction of ADAMTS13 M domain (PDB: 6QIG) with the catalytic Glu225 residue (blue) replacing the Gln225 in the crystal structure for illustrative purposes. Active site residues His224, His228, and His234 (blue) coordinate the zinc ion (green sphere). Most active site residues are part of the α4 helix (other helix residues shown in gray). Other M domain regions with evidence of increased deuterium uptake are shown here, namely the α5 helix and the α2 helix (dark gray). (B) M domain active site residues shown in blue as in panel A, with important areas for substrate binding depicted in light green. Specifically highlighted are the S1 pocket (thought to bind VWF residue Tyr1605), S1ʹ pocket (thought to bind VWF Met1606), and S3 pocket (thought to bind VWF L1603). Leu232 is immediately C-terminal to the α4 helix. (C-E) Residues with increased deuterium uptake in the presence of scFv3-1, scFv4-16, and scFv4-20, respectively, involving the active site (magenta) and other regions of the M domain with increased deuterium uptake (red), are illustrated.
ADAMTS13 M domain residues exhibit a conformational change in the presence of antispacer domain inhibitors. (A) Depiction of ADAMTS13 M domain (PDB: 6QIG) with the catalytic Glu225 residue (blue) replacing the Gln225 in the crystal structure for illustrative purposes. Active site residues His224, His228, and His234 (blue) coordinate the zinc ion (green sphere). Most active site residues are part of the α4 helix (other helix residues shown in gray). Other M domain regions with evidence of increased deuterium uptake are shown here, namely the α5 helix and the α2 helix (dark gray). (B) M domain active site residues shown in blue as in panel A, with important areas for substrate binding depicted in light green. Specifically highlighted are the S1 pocket (thought to bind VWF residue Tyr1605), S1ʹ pocket (thought to bind VWF Met1606), and S3 pocket (thought to bind VWF L1603). Leu232 is immediately C-terminal to the α4 helix. (C-E) Residues with increased deuterium uptake in the presence of scFv3-1, scFv4-16, and scFv4-20, respectively, involving the active site (magenta) and other regions of the M domain with increased deuterium uptake (red), are illustrated.
Discussion
Autoantibodies against ADAMTS13 play a central role in the pathophysiology of iTTP. ADAMTS13 likely is intrinsically disordered, which emphasizes the need for functional studies to better understand the mechanism of action of ADAMTS13 and how inhibitors render it nonfunctional.40 In this study, we explored whether antispacer antibodies primarily affect substrate concentration–related effects on enzymatic function, or whether they affect activity by some other mechanisms. We used a panel of human monoclonal inhibitory scFvs that bind to the spacer domain, the major target of most inhibitory antibodies in iTTP.8,9,11-13,41 We found that, in all temperature and pH conditions tested with both recombinant and plasma ADAMTS13, that there was a significant effect on Vmax by each inhibitor when the enzyme was titrated with a fluorogenic surrogate VWF73 substrate. In contrast, effects on K0.5 were not nearly as robust, with 2 of the 3 tested inhibitors (scFv4-20 and scFv4-16) showing little to no effect on this parameter, whereas scFv3-1 may have had some effect on K0.5. We were unable to use acidic concentrations closer to physiologic pH because enzyme activity is too low under those circumstances and does not allow for quantitative assessment of its activity. However, by using a proton concentration fivefold lower than standard (pH 6.0 vs pH 6.31), we did not see any observable difference in the apparent mechanism. Experiments at physiologic temperature (37°C) also revealed similar results.
To our knowledge, this study is the first to explore the effects of human monoclonal anti-ADAMTS13 antibodies on Vmax and K0.5. Results suggest that inhibition of ADAMTS13 in iTTP may be achieved by altering the rate of catalytic turnover without significantly affecting substrate concentration effects on activity, although the inhibitors bind to an important substrate-binding exosite.11,12,24,41-44 Although ADAMTS13 binds VWF73 at several distinct sites, the results are unexpected because the epitope has clearly been implicated in substrate binding and ADAMTS13-mediated cleavage.45,46 In fact, small peptides that mimic part of the scFv4-20 binding site have previously been shown to inhibit ADAMTS13-mediated cleavage of VWF73, presumably by acting as a competitive inhibitor.44 In this context, the results of this study differ from expectations but does suggest an additional mechanism underlying autoantibody-mediated inhibition of plasma ADAMTS13 activity.
We used HX-MS to explore the mechanism of the effect on catalytic turnover by these antispacer antibodies. Our hypothesis was that inhibitory antibodies that bind the spacer domain may allosterically affect the conformation of the catalytic site in the metalloprotease domain. With all 3 inhibitory antibodies, we observed increased deuterium uptake in α4 helix that includes 3 of the 4 residues that make up the active site of the enzyme (Figure 5). This suggests that antispacer antibodies may prevent VWF cleavage by disrupting the secondary structure of the region containing His228, which may affect the ability of the metalloprotease to coordinate the important zinc ion at the active site. It is possible that the conformation of this region in the presence of inhibitory antibodies also represents the conformation of ADAMTS13 in its latent state, because we have recently found that, when compared with MDTCS, full-length ADAMTS13 also demonstrates increased deuterium uptake in this region.47 Ongoing work will explore how antispacer antibodies may affect the binding of zinc to the metalloprotease domain to corroborate this hypothesis.
Notably, our data also suggest that increased deuterium uptake occurs immediately distal to the α4 helix in the presence of all 3 antispacer antibodies tested. This region includes the L232 involved in the S3 pocket that helps bind the VWF residue L1603; previous work has shown that this residue has an essential role in ADAMTS13-mediated VWF cleavage.48,49 Our data suggest that antispacer antibodies, as a class, may affect the ability of the enzyme to bind to VWF at L1603. Such an effect may contribute to the increase in K0.5 seen with some of the inhibitors under different experimental conditions, particularly with scFv3-1. Future work by our group will help to determine how disruption of this interaction may contribute to the mechanism of action of ADAMTS13 inhibitors.
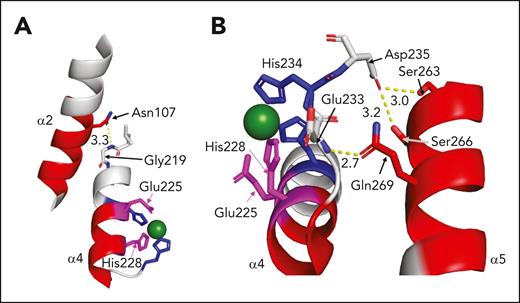
The residues shown to have increased uptake of deuterium in the presence of antispacer inhibitors may be involved in interactions elsewhere in the protein. The side chain of N107 in the α2 helix forms a hydrogen bond with the backbone of G219 in the α4 helix (Figure 6A).50 Because N107 is part of a peptide region with increased deuterium uptake in the presence of the inhibitor scFv3-1, it is reasonable to think that disruption of this interaction may have global effects on the tertiary structure of the M domain. The inhibitors scFv3-1 and scFv4-20 also affected deuterium uptake in the α5 helix (S263-A276), specifically in peptide fragments containing residues W262 to S272 (Figure 4; supplemental Figure 2). A hydrogen bond between the Q269 side chain and the backbone of E233, as well as side chain ionic interactions between α5 helix residues S263 and S266 with the side chain of D235, appear to be involved in the tertiary structure of the protein between the α4 and α5 helices (Figure 6B).50 Data from cross-linking mass spectrometry experiments suggest that there is an interaction between S266 on ADAMTS13 and the VWF residue Y1605, an enticing connection that may further explain how inhibitors affect enzymatic function.40 Although the antispacer domain inhibitors bind to similar regions of the protein with significant epitope overlap, their allosteric effects and, as such, their mechanisms of inhibition, may differ to some extent. Nonetheless, increased deuterium uptake in the active site itself was seen in the presence of all inhibitors tested, and it is possible that this is the central inhibitory mechanism of anti-ADAMTS13 antibodies.
Potential contacts between the M regions with increased deuterium uptake (magenta: active site residues; red: other M residues) and the other regions of the M domain in the presence of scFv3-1. Zinc ion is depicted as a green sphere. (A) A polar contact between the side chain of Asn107 in the α2 helix and the backbone of Gly219 in the α4 helix, which contains 3 of the 4 residues in the active site, is illustrated. (B) Polar contacts between the side chain of Gln269 in the α5 helix and the backbone of Glu233 immediately N-terminal to His234, which helps coordinate the zinc ion; and the side chains of α5 residues S263 and S266 with the side chain of Asp235 immediately C-terminal to His234, are illustrated.
Potential contacts between the M regions with increased deuterium uptake (magenta: active site residues; red: other M residues) and the other regions of the M domain in the presence of scFv3-1. Zinc ion is depicted as a green sphere. (A) A polar contact between the side chain of Asn107 in the α2 helix and the backbone of Gly219 in the α4 helix, which contains 3 of the 4 residues in the active site, is illustrated. (B) Polar contacts between the side chain of Gln269 in the α5 helix and the backbone of Glu233 immediately N-terminal to His234, which helps coordinate the zinc ion; and the side chains of α5 residues S263 and S266 with the side chain of Asp235 immediately C-terminal to His234, are illustrated.
These results suggest that conformational changes at the active site itself in the presence of antispacer domain inhibitors contribute both to ADAMTS13 inhibition as well as its latent state. Alternatively, it is hypothesized that a so-called “gatekeeper triad,” formed by interaction of R193, D217, and D252, prevents the VWF residue M1606 from binding to the S1ʹ pocket (ADAMTS13 residues D252-P256) in the latent state of ADAMTS13.48-50 This triad was structurally identified in the context of a recombinant ADAMTS13 with the active site E225 substituted with glutamine, with an inhibitory mouse antihuman anti-M scFv (3H9) bound to it. It is possible that the triad is a feature of inhibition, latency, or both. Our data do not show significant differences in deuterium uptake in these residues, but this in no way excludes the possibility of multiple mechanisms at play in the functioning and inhibition of ADAMTS13; rather, multiple mechanisms may be involved. Future work will need to consider all these hypotheses when exploring the pathophysiology of iTTP.
Studies focused on the manner(s) in which the enzyme can be rendered nonfunctional are needed to develop better ways of preserving and recovering ADAMTS13 function. The tools described here in this study will be helpful to test the mechanisms of action of human monoclonal antibodies against ADAMTS13 from patients with iTTP as they are identified. This work may also contribute to the development of ADAMTS13 constructs not affected by anti-ADAMTS13 inhibitors, an avenue being explored by our group and others.42,51 Our ongoing studies are focused on conformational effects on ADAMTS13 by antibodies using both structural techniques as well as binding assays. Although scFvs are powerful tools to evaluate Fab-mediated effects of antibodies, future work will explore how intact IgG affects inhibition, possibly by avidity-related mechanisms. These studies are probing the effects of antibodies on ADAMTS13 and VWF, as well as their effects on the binding affinity of other antibodies. Studies such as this will ideally lead to exploration of molecular targets on ADAMTS13 that prevent antibodies from binding to it, reverse antibody binding, or, possibly, optimize its function when used to treat patients with iTTP.
Acknowledgments
The authors thank Lucy Zheng of Bryn Mawr College for artwork in the visual abstract.
The study was supported in part by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (HL126724, HL144552, HL157975-01A1 [X.L.Z.] and HL163471-01 [K.H.]), a mentored research award from the Hemostasis and Thrombosis Research Society (K.H.), and a career development award from the American Heart Association (K.H.).
Authorship
Contribution: K.H., C.M., and X.L.Z. designed the study, analyzed the results, and wrote the manuscript; S.L. and L.M. performed experiments and data analysis, and revised the manuscript; D.L.S. provided critical reagents and revised manuscript; and all authors approved the final version of the manuscript for submission.
Conflict-of-interest disclosure: X.L.Z. is a speaker and/or consultant for Alexion, Sanofi, and Takeda and is the cofounder of Clotsolution. The remaining authors declare no competing financial interests.
Correspondence: Konstantine Halkidis, Division of Hematologic Malignancies and Cellular Therapeutics, Department of Internal Medicine, The University of Kansas Medical Center, 3901 Rainbow Blvd, Kansas City, KS 66160; e-mail: khalkidis@kumc.edu; and X. Long Zheng, Department of Pathology and Laboratory Medicine, The University of Kansas Medical Center, 3901 Rainbow Blvd, Kansas City, KS 66160;.
References
Author notes
∗K.H. and C.M. contributed equally to this study.
Data are available on request from the corresponding authors, Konstantine Halkidis (khalkidis@kumc.edu) and X. Long Zheng (xzheng2@kumc.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal