Key Points
BRD4 protects DLBCL cells from ferroptosis by positively regulating the expression of FSP1.
BET inhibitors increase the susceptibility of GCB-DLBCL cells to ferroptosis and thus promote the toxicity of DMF both in vitro and in vivo.
Abstract
Diffuse large B-cell lymphoma (DLBCL), the most common form of non-Hodgkin lymphoma, is characterized by an aggressive clinical course. In approximately one-third of patients with DLBCL, first-line multiagent immunochemotherapy fails to produce a durable response. Molecular heterogeneity and apoptosis resistance pose major therapeutic challenges in DLBCL treatment. To circumvent apoptosis resistance, the induction of ferroptosis might represent a promising strategy for lymphoma therapy. In this study, a compound library, targeting epigenetic modulators, was screened to identify ferroptosis-sensitizing drugs. Strikingly, bromodomain and extra-terminal domain (BET) inhibitors sensitized cells of the germinal center B-cell–like (GCB) subtype of DLBCL to ferroptosis induction and the combination of BET inhibitors with ferroptosis inducers, such as dimethyl fumarate or RSL3, synergized in the killing of DLBCL cells in vitro and in vivo. On the molecular level, the BET protein BRD4 was found to be an essential regulator of ferroptosis suppressor protein 1 expression and thus to protect GCB-DLBCL cells from ferroptosis. Collectively, we identified and characterized BRD4 as an important player in ferroptosis suppression in GCB-DLBCL and provide a rationale for the combination of BET inhibitors with ferroptosis-inducing agents as a novel therapeutic approach for DLBCL treatment.
Introduction
Globally, non-Hodgkin lymphoma represents one of the most prevalent cancers with increasing incidence.1 Among non-Hodgkin lymphoma, with 30% to 40% of cases diffuse large B-cell lymphoma (DLBCL) constitutes the most frequent entity in adults and is characterized by an aggressive clinical course.1,2 DLBCL is a heterogeneous diagnostic category regarding clinical presentation, genetic aberrations, and therapy response. Using gene expression profiling, DLBCL can be subdivided at the molecular level into 2 major subtypes, ie, germinal center B-cell–like (GCB) and activated B-cell–like (ABC) DLBCL.3-5 Clustering of concomitant genetic alterations allowed the classification of up to 7 different DLBCL subtypes, improving the prediction of the response to first-line and targeted therapies.6-8 Despite these advances in the molecular characterization of DLBCL subtypes, the multiagent chemotherapy that includes CHOP (cyclophosphamide, hydroxydaunorubicin, oncovin and prednisone) and is supplemented by the anti-CD20 antibody rituximab (R) still represents the first-line standard-of-care treatment for all DLBCL subtypes.9-11 While approximately two-thirds of patients with DLBCL show a durable response to R-CHOP, the remaining patients only respond partially or relapse after an initial response.1,9-11
Resistance of lymphoma cells toward apoptotic cell death is one of the major reasons underlying the failure of multiagent chemotherapy.12 The molecular mechanisms governing apoptosis resistance in DLBCL are diverse and include, for instance, the inactivation of the tumor suppressor p53 by mutations and deletions of TP53 or amplification of MDM2, overexpression of BCL6, or mutations of ATM and CREBBP.12,13 Furthermore, enhanced expression of prosurvival members of the BCL2 family, such as BCL2, MCL1, BCL-x, or BFL-1, is observed in the majority of biopsy from patients with DLBCL and is thought to contribute strongly to apoptosis resistance.12,13
In contrast to apoptosis, it has been demonstrated that DLBCL cells are highly vulnerable to ferroptosis, an iron-dependent form of cell death that exhibits unique features, which clearly distinguish it from other types of regulated cell death.14,15 The hallmark of ferroptosis is the iron-catalyzed oxidation of polyunsaturated fatty acid chains, which eventually results in the loss of cellular membrane integrity.16-19 To prevent a deleterious radical chain reaction within cellular membranes, lipid peroxides are removed rapidly by various detoxifying mechanisms. Two major enzymes, the glutathione peroxidase 4 (GPX4) and the ferroptosis suppressor protein 1 (FSP1), are crucial for the suppression of excessive lipid peroxidation.15,20,21 Whereas GPX4 catalyzes the reduction of lipid peroxides in cellular membranes to the corresponding alcohols in a glutathione-dependent manner, FSP1 uses nicotinamide adenine dinucleotide phosphate (NAD[P]H) to reduce its substrates, such as coenzyme Q10, vitamin K, and α-tocopherol, which in turn are able to scavenge lipid peroxyl radicals.15,20-22 While pharmacological inhibitors of GPX4 and FSP1 have not yet entered clinical applications, we have recently identified the Food and Drug administration–approved drug dimethyl fumarate (DMF), which is used in the treatment of relapsing-remitting multiple sclerosis and moderate-to-severe psoriasis, as potent ferroptosis inducer (FIN) with a broad antilymphoma effect against DLBCL cells.14 Mechanistically, the electrophile DMF depletes the cellular glutathione pool in an irreversible process, termed succination, thus impairing GPX4 activity and promoting lipid peroxidation.
The mammalian bromodomain and extra-terminal domain (BET) protein family comprises 4 conserved members, the bromodomain-containing proteins 2 to 4 (BRD2-4) and the testis-specific BRDT.23,24 Each BET family member harbors 2 amino-terminal bromodomains via which it recognizes acetylated lysine residues in transcription regulatory proteins and histones, and thus contributes to genome stability and transcriptional control. By targeting their bromodomains, BET inhibitors (BETis) reduce the presence of the BET proteins in enhancer and promoter regions. The anticancer activity of BETis in hematologic malignancies, for example acute myeloid leukemia, has been attributed to the downregulation of oncogenes, such as MYC.25,26 The best studied BET protein BRD4 promotes transcription on various levels. By interacting with acetylated histones as well as transcription factors and by simultaneously recruiting the mediator complex, BRD4 serves as a platform for diverse transcriptional regulators and bridges super-enhancer and promoter regions, thus stabilizing the transcriptional machinery.27 Furthermore, BRD4 promotes the activity of RNA-polymerase II by recruiting the positive transcription elongation factor b (P-TEFb) complex.27-31
In this study, we used a chemical compound library targeting a variety of epigenetic regulators to screen for drugs that promote ferroptosis induction in DLBCL cells. We identified BRD4 as a promising target, because it controlled FSP1 expression and protected DLBCL cells from ferroptosis. Accordingly, BETis synergized with FINs, such as DMF, in the killing of DLBCL cells in vitro and in vivo and thus might represent a novel treatment option for this malignancy.
Materials and methods
Cell culture, transfection, lentiviral transduction, and survival assays
Protocols are provided in the supplemental Materials and Methods; available on the Blood website.
Quantification of cellular glutathione levels
Levels of reduced and oxidized glutathione in cellular lysates were quantified using the GSH/GSSG-Glo Assay (Promega) according to the manufacturer’s protocol.
Cell lysis, immunoblotting, and chromatin immunoprecipitation
Protocols are provided in the supplemental Materials and Methods.
Gene expression profiling, gene set enrichment analysis and quantitative polymerase chain reaction
Protocols are provided in the supplemental Materials and Methods.
Mass spectrometry for metabolomics, analysis of lipid peroxidation, and xenograft mouse models
Protocols are provided in the supplemental Materials and Methods.
Quantification of malondialdehyde
Malondialdehyde levels in tumor samples were quantified using the thiobarbituric acid reactive substances assay (Cayman chemical) according to the manufacturer’s instructions.
Informed consent to donation of lymphoma samples for the derivation of murine patient-derived xenografts was obtained from patients according to the Declaration of Helsinki.
Results
BRD4 inhibition promotes lipid peroxidation
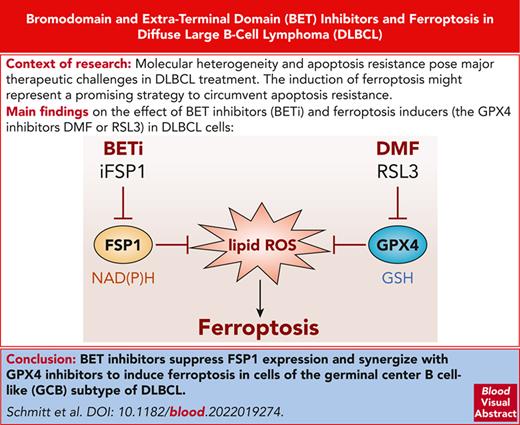
To identify novel FINs or substances that amplify DMF-dependent ferroptosis, we treated the DLBCL cell line DOHH2 which is highly susceptible to ferroptosis induction with ∼140 epigenetic modulators alone or in combination with DMF and assessed the extent of lipid peroxidation by flow cytometry (Figure 1A). Although the majority of the library compounds did not promote or even protected the cells from DMF-induced lipid peroxidation, several BETis, such as bromosporine, RVX-208, UMB-32, BI-2536, AZD5153, I-BET151, and JQ1, amplified the DMF-mediated increase in lipid peroxides (Figure 1B; supplemental Figure 1). Treatment of DOHH2 cells with different BETis (JQ1, PFI-1, I-BET151, and I-BET762) alone did not provoke lipid peroxidation but enhanced the DMF-induced accumulation of lipid peroxides in a dose-dependent manner (Figure 1C; supplemental Figure 2A-B). To determine whether this BETi-dependent sensitization can be observed in different molecular DLBCL subtypes, we analyzed lipid peroxidation after BET inhibition and concomitant DMF treatment in a set of GCB- and ABC-DLBCL cell lines. Although all investigated GCB-DLBCL cell lines exhibited an increase in DMF-induced lipid peroxidation after BETi treatment, none of the 5 ABC-DLBCL cell lines were sensitized by BET inhibition (Figure 1D; supplemental Figure 3A-C). To investigate whether BET inhibition promotes lipid peroxidation in GCB-DLBCL cells not only in response to DMF but also to other FINs, we treated various GCB-DLBCL cell lines with the GPX4-inhibitor RSL3 alone or in combination with JQ1 and I-BET151. Both BETis significantly increased RSL3-dependent lipid peroxidation, suggesting that BET inhibition generally sensitizes GCB-DLBCL cells to the accumulation of lipid peroxides (Figure 1E; supplemental Figure 4A-B). Because the majority of BETis included in the compound library target the bromodomains of BRD4 and because UMB-32 as well as AZD5153 are considered BRD4-specific inhibitors, we speculated that BRD4 plays a central role in the protection from lipid peroxidation in DLBCL cells. Indeed, short-hairpin RNA (shRNA)–mediated silencing of BRD4 expression enhanced DMF-dependent lipid peroxidation (Figure 1F; supplemental Figure 4C). In conclusion, we demonstrate that targeting of BRD4 with BET inhibitors sensitizes GCB-DLBCL cells to DMF-induced lipid peroxidation.
BET inhibition amplifies DMF-induced lipid peroxidation in DLBCL. (A) Experimental setup of the compound library screen comprising epigenetic modulators used for the identification of ferroptosis-sensitizing agents. (B) DOHH2 cells were treated with the indicated compounds for 24 hours and subsequently incubated with DMF for 2 hours. Lipid peroxidation was quantified by flow cytometry using the oxidation-sensitive fluorescent probe BODIPY C11 and normalized to the solvent control. (C) DOHH2 cells were treated with the indicated BETis for 24 hours and DMF for an additional 2 hours before analyzing lipid peroxidation by flow cytometry. (D) The indicated GCB-DLBCL cell lines were treated with either JQ1, DMF, or the combination of both compounds. Lipid peroxidation was assessed using flow cytometry. (E) Quantification of lipid peroxidation in DOHH2 cells that were treated with JQ1 or I-BET151 for 24 hours and RSL3 for another 2 hours. The mean fluorescence intensity (MFI) of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (F) Lipid peroxidation of DMF-treated control and BRD4-silenced DOHH2 cells was analyzed using flow cytometry. Error bars correspond to the mean ± standard deviations (SD). Data are representative of at least 3 (C-E) or 2 (F) independent experiments. ∗P < .05; ∗∗∗P < .005.
BET inhibition amplifies DMF-induced lipid peroxidation in DLBCL. (A) Experimental setup of the compound library screen comprising epigenetic modulators used for the identification of ferroptosis-sensitizing agents. (B) DOHH2 cells were treated with the indicated compounds for 24 hours and subsequently incubated with DMF for 2 hours. Lipid peroxidation was quantified by flow cytometry using the oxidation-sensitive fluorescent probe BODIPY C11 and normalized to the solvent control. (C) DOHH2 cells were treated with the indicated BETis for 24 hours and DMF for an additional 2 hours before analyzing lipid peroxidation by flow cytometry. (D) The indicated GCB-DLBCL cell lines were treated with either JQ1, DMF, or the combination of both compounds. Lipid peroxidation was assessed using flow cytometry. (E) Quantification of lipid peroxidation in DOHH2 cells that were treated with JQ1 or I-BET151 for 24 hours and RSL3 for another 2 hours. The mean fluorescence intensity (MFI) of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (F) Lipid peroxidation of DMF-treated control and BRD4-silenced DOHH2 cells was analyzed using flow cytometry. Error bars correspond to the mean ± standard deviations (SD). Data are representative of at least 3 (C-E) or 2 (F) independent experiments. ∗P < .05; ∗∗∗P < .005.
BETis and FINs synergistically induce ferroptosis
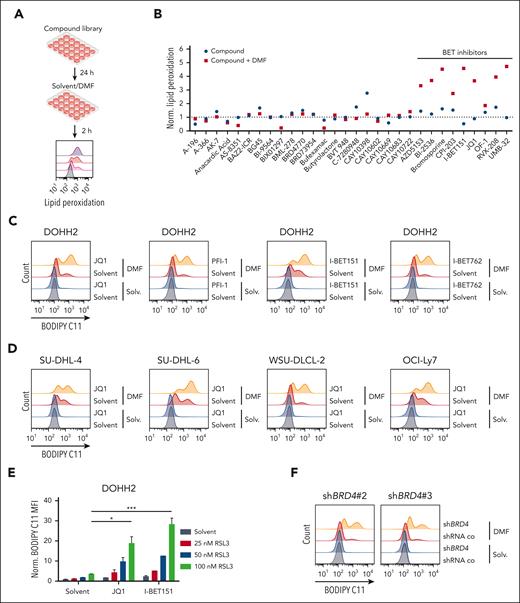
To investigate whether BET inhibition promotes the cytotoxic effect of FINs, we treated 3 GCB-DLBCL cell lines (DOHH2, SU-DHL-6, and WSU-DLCL-2) with DMF, JQ1, or the combination of both inhibitors. Strikingly, the combinatorial treatment reduced the cell numbers rapidly and synergistically (Figure 2A; supplemental Figure 5A-B). Similarly, cotreatment with I-BET151 also amplified the cytotoxicity of DMF in GCB-DLBCL cell lines (supplemental Figure 6). In accordance with the increased sensitivity of BRD4-silenced cells to DMF-mediated lipid peroxidation, reduced BRD4 expression rendered DOHH2 cells more susceptible to DMF-induced toxicity (Figure 2B; supplemental Figure 4C). In contrast to GCB-DLBCL, the combination treatment with DMF and JQ1 did not result in synergistic cytotoxicity in ABC-DLBCL (supplemental Figure 7A-B). To verify that the combination treatment of BETis with DMF indeed induced ferroptosis in GCB-DLBCL, we assessed the impact of iron chelators (deferoxamine), lipophilic antioxidants (α-tocopherol and ferrostatin-1), and the glutathione precursor N-acetyl-L-cysteine on lipid peroxidation and cell death. Indeed, all substances were able to prevent lipid peroxidation triggered by the combinatorial BETi/DMF treatment (supplemental Figure 8A). Accordingly, antioxidant treatment and iron chelation restored cell viability to the level of the single BETi treatment (JQ1 or I-BET151) in several GCB-DLBCL cell lines, indicating that BET inhibition indeed promoted DMF-induced ferroptosis, whereas the cytotoxicity of the BETi single treatment was not associated with ferroptosis (Figure 2C; supplemental Figure 8B-C). In contrast, the broad-spectrum caspase inhibitor Q-VD-OPh was unable to mitigate the cytotoxicity induced by the combined treatment with BETis and DMF (Figure 2C; supplemental Figure 8B). As the combination of JQ1 and the GPX4 inhibitor RSL3 also reduced cell numbers synergistically, we concluded that BETis possess the general capacity of sensitizing GCB-DLBCL cells to ferroptosis induction (Figure 2D; supplemental Figure 9A).
BET inhibition sensitizes DLBCL cell lines to ferroptosis induction. (A) DOHH2 and SU-DHL-6 cells were treated with DMF alone (left panels) or in combination with JQ1 (right panels), as indicated. Cells were treated with JQ1 for 24 hours before incubation with DMF for another 24 hours. Cell numbers were determined and the combination treatments were normalized to the respective DMF single treatment. The CI for 20 μM DMF and 0.25 μM JQ1 in DOHH2 and SU-DHL-6 cells is 0.26 and 0.46, respectively. (B) The survival of BRD4-silenced DOHH2 cells in response to daily DMF treatment was determined after 3 days and normalized to control cells transduced with a nontargeting shRNA. (C) DOHH2 cells were treated with 0.25 μM JQ1 or 0.25 μM I-BET151 for 24 hours before adding 10 μM DMF alone or in combination with 10 μM ferrostatin-1 (Fer-1), 100 μM α-tocopherol (α-Toco), 100 μM deferoxamine (DFX), 25 μM N-acetyl-L-cysteine (NAC), and 10 μM Q-VD-OPh for another 24 hours, as indicated. After 48 hours, cell numbers were determined. Dotted line marks cell survival upon single treatment with BETi. (D) DOHH2 and SU-DHL-6 cell were treated with JQ1 and RSL3 either alone or in combination for 48 hours before quantifying cell survival. The combination treatments were normalized to the respective RSL3 single treatment. The CI in DOHH2 cells was 0.16 (for 0.05 μM RSL3 and 0.1 μM JQ1) and in SU-DHL-6 cells 0.13 (for 0.01 μM RSL3 and 0.1 μM JQ1). (E) DOHH2, SU-DHL-4, and SU-DHL-6 cells were treated with solvent, LDC067, DMF, or the indicated combinations. Lipid peroxidation was analyzed by flow cytometry. (F) DOHH2 and SU-DHL-6 cells were treated with DMF (left panels) alone or in combination with LDC067 (right panels), as indicated. Cell survival was quantified after 48 hours of treatment. The CI in DOHH2 cells was 0.60 (for 10 μM DMF and 5 μM LDC067) and in SU-DHL-6 cells 0.51 (for 20 μM DMF and 2.5 μM LDC067). Error bars correspond to the mean ± SD. Data are representative of at least 3 (A,D-F) or 2 (B-C) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. CI, combination index.
BET inhibition sensitizes DLBCL cell lines to ferroptosis induction. (A) DOHH2 and SU-DHL-6 cells were treated with DMF alone (left panels) or in combination with JQ1 (right panels), as indicated. Cells were treated with JQ1 for 24 hours before incubation with DMF for another 24 hours. Cell numbers were determined and the combination treatments were normalized to the respective DMF single treatment. The CI for 20 μM DMF and 0.25 μM JQ1 in DOHH2 and SU-DHL-6 cells is 0.26 and 0.46, respectively. (B) The survival of BRD4-silenced DOHH2 cells in response to daily DMF treatment was determined after 3 days and normalized to control cells transduced with a nontargeting shRNA. (C) DOHH2 cells were treated with 0.25 μM JQ1 or 0.25 μM I-BET151 for 24 hours before adding 10 μM DMF alone or in combination with 10 μM ferrostatin-1 (Fer-1), 100 μM α-tocopherol (α-Toco), 100 μM deferoxamine (DFX), 25 μM N-acetyl-L-cysteine (NAC), and 10 μM Q-VD-OPh for another 24 hours, as indicated. After 48 hours, cell numbers were determined. Dotted line marks cell survival upon single treatment with BETi. (D) DOHH2 and SU-DHL-6 cell were treated with JQ1 and RSL3 either alone or in combination for 48 hours before quantifying cell survival. The combination treatments were normalized to the respective RSL3 single treatment. The CI in DOHH2 cells was 0.16 (for 0.05 μM RSL3 and 0.1 μM JQ1) and in SU-DHL-6 cells 0.13 (for 0.01 μM RSL3 and 0.1 μM JQ1). (E) DOHH2, SU-DHL-4, and SU-DHL-6 cells were treated with solvent, LDC067, DMF, or the indicated combinations. Lipid peroxidation was analyzed by flow cytometry. (F) DOHH2 and SU-DHL-6 cells were treated with DMF (left panels) alone or in combination with LDC067 (right panels), as indicated. Cell survival was quantified after 48 hours of treatment. The CI in DOHH2 cells was 0.60 (for 10 μM DMF and 5 μM LDC067) and in SU-DHL-6 cells 0.51 (for 20 μM DMF and 2.5 μM LDC067). Error bars correspond to the mean ± SD. Data are representative of at least 3 (A,D-F) or 2 (B-C) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. CI, combination index.
BRD4 has been implicated in diverse cellular processes that include not only the regulation of chromatin remodeling and the maintenance of DNA integrity but also the control of transcription via recruitment of the P-TEFb complex. The P-TEFb complex comprises the cyclin-dependent kinase 9 (CDK9), which phosphorylates transcription-pausing factors and the carboxy-terminal domain of RNA-polymerase II, thus promoting transcriptional activity.27-31 To investigate whether the P-TEFb complex and particularly CDK9 participate in ferroptosis protection, we treated various GCB-DLBCL cell lines with the selective CDK9 inhibitor LDC067 and assessed both lipid peroxidation as well as survival in response to DMF treatment. The CDK9 inhibitor by itself did not provoke an increase in lipid peroxidation, but, strikingly, promoted the DMF-induced effects (Figure 2E). Accordingly, combinatorial treatment with LDC067 and DMF resulted in the synergistic killing of GCB-DLBCL cells (Figure 2F; supplemental Figure 9B). Taken together, we demonstrate that transcriptional activity induced by the BRD4/P-TEFb axis is crucial for the prevention of excessive lipid peroxidation in GCB-DLBCL cells.
BET proteins control the expression of ferroptosis-associated genes
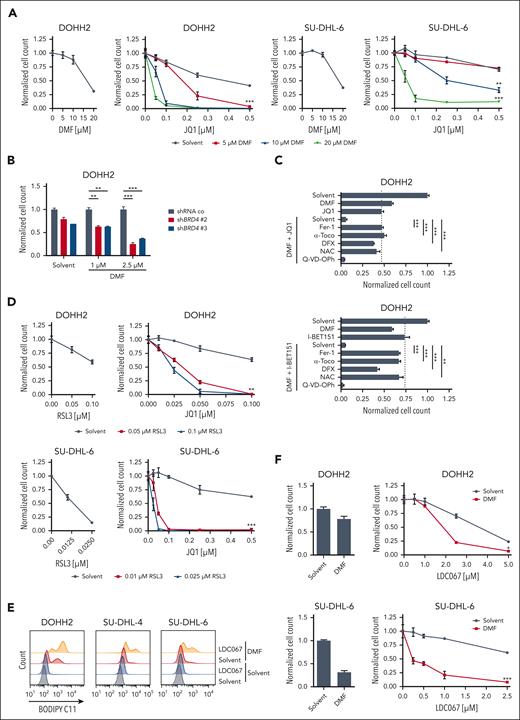
As inhibition of both BRD4 and CDK9 increases the ferroptosis susceptibility of GCB-DLBCL cells, we hypothesized that transcriptional changes are responsible for the observed sensitization to ferroptotic cell death. To identify ferroptosis-related genes that are altered in their expression upon BET inhibition, we treated DOHH2 cells for 12 or 24 hours with 0.25 μM JQ1 and analyzed gene expression changes by RNA-sequencing. Using stringent selection criteria, we identified 51 upregulated and 86 downregulated genes in BETi-treated cells (Figure 3A; supplemental Figure 10A-B; supplemental Table 6). Gene set enrichment and gene ontology analyses revealed that BET inhibition affected the expression of genes controlling B-cell receptor–mediated signaling, innate immune responses, amino acid transmembrane transport, and the respiratory burst (supplemental Figure 11-12; supplemental Table 7). Interestingly, in addition to the previously described BRD4 target gene TLR10, also AIFM2 and SLC7A11, which encode proteins essential for the suppression of ferroptosis, were downregulated in DOHH2 cells in response to JQ1 treatment (Figure 3A-B).32 In contrast, the expression of other ferroptosis-associated genes that are involved in iron metabolism (FTL and FTH1), glutathione biosynthesis (GCLC and GCLM), and the detoxification of lipid peroxides (GPX4) was not affected by BET inhibition (Figure 3B; supplemental Figure 13A). As a subunit of the system xc- transporter, SLC7A11 mediates the import of extracellular cystine which is subsequently reduced to cysteine and serves as substrate for glutathione biosynthesis.33 In all GCB-DLBCL cell lines investigated, JQ1 treatment reduced SLC7A11 transcript levels in a dose-dependent manner (Figure 3B-C; supplemental Figure 13A). Not only JQ1 but also I-BET151, I-BET762, and PFI-1 efficiently impaired SLC7A11 mRNA expression in DOHH2 cells (supplemental Figure 13B). Furthermore, SLC7A11 protein levels were reduced in several GCB-DLBCL cell lines in response to BET inhibition (Figure 3D).
BET inhibitors control the expression of ferroptosis-associated genes in DLBCL. (A) Heatmap of genes differentially expressed in DOHH2 cells treated with 0.25 μM JQ1 for 12 or 24 hours compared to the solvent control. Gene expression changes are depicted according to the color scale. (B) DOHH2 cells were treated with 0.25 μM JQ1 for 12 or 24 hours. Transcript levels of the indicated genes were quantified by qPCR. Expression in JQ1-treated cells was normalized to the respective solvent control. SDHA served as reference gene. (C) SLC7A11 mRNA levels of solvent- or JQ1-treated GCB-DLBCL cell lines were determined by qPCR. SDHA served as reference gene. (D) The indicated GCB-DLBCL cell lines were incubated with 0.25 μM JQ1 or I-BET151 for 48 hours. SLC7A11 protein levels were visualized by immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as loading control. (E) Reduced GSH was quantified in the indicated GCB-DLBCL cell lines upon treatment with solvent, JQ1 or I-BET151 for 24 hours. GSH concentrations were normalized to the respective solvent control. (F) DOHH2 cells were treated with 0.25 μM JQ1 or I-BET151 alone or in combination with different concentrations of DMF. GSH levels were quantified and normalized to the solvent control. (G) The GCB-DLBCL cell line DOHH2 and the ABC-DLBCL cell line U2932 were treated with 0.25 μM JQ1 alone or in combination with the indicated concentrations of DMF. GSH levels were quantified and normalized to the respective DMF-treated controls. Error bars correspond to the mean ± SD. Data are representative of at least 3 (B-C,E-F) or 2 (D,G) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. GSH, glutathione; mRNA, messenger RNA; n.s., not significant; qPCR, quantitative polymerase chain reaction.
BET inhibitors control the expression of ferroptosis-associated genes in DLBCL. (A) Heatmap of genes differentially expressed in DOHH2 cells treated with 0.25 μM JQ1 for 12 or 24 hours compared to the solvent control. Gene expression changes are depicted according to the color scale. (B) DOHH2 cells were treated with 0.25 μM JQ1 for 12 or 24 hours. Transcript levels of the indicated genes were quantified by qPCR. Expression in JQ1-treated cells was normalized to the respective solvent control. SDHA served as reference gene. (C) SLC7A11 mRNA levels of solvent- or JQ1-treated GCB-DLBCL cell lines were determined by qPCR. SDHA served as reference gene. (D) The indicated GCB-DLBCL cell lines were incubated with 0.25 μM JQ1 or I-BET151 for 48 hours. SLC7A11 protein levels were visualized by immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as loading control. (E) Reduced GSH was quantified in the indicated GCB-DLBCL cell lines upon treatment with solvent, JQ1 or I-BET151 for 24 hours. GSH concentrations were normalized to the respective solvent control. (F) DOHH2 cells were treated with 0.25 μM JQ1 or I-BET151 alone or in combination with different concentrations of DMF. GSH levels were quantified and normalized to the solvent control. (G) The GCB-DLBCL cell line DOHH2 and the ABC-DLBCL cell line U2932 were treated with 0.25 μM JQ1 alone or in combination with the indicated concentrations of DMF. GSH levels were quantified and normalized to the respective DMF-treated controls. Error bars correspond to the mean ± SD. Data are representative of at least 3 (B-C,E-F) or 2 (D,G) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. GSH, glutathione; mRNA, messenger RNA; n.s., not significant; qPCR, quantitative polymerase chain reaction.
To analyze whether BET inhibition affects the cellular antioxidant capacity required to restrict the deleterious effects of ongoing lipid peroxidation, we quantified the intracellular levels of the GPX4 substrate glutathione, the FSP1 cofactor NAD(P)H and mevalonate, a precursor of coenzyme Q10, by mass spectometry. While BET inhibition did not significantly alter the levels of NADH, NADPH, and mevalonate, BETi treatment moderately decreased the amount of reduced glutathione (Figure 3E; supplemental Figure 14A-C). In contrast to the DMF-dependent depletion of reduced glutathione, the effect of BET inhibition was rather modest (Figure 3F). However, BETi treatment cooperated with DMF in reducing the intracellular glutathione pool in GCB-DLBCL cell lines (Figure 3F-G; supplemental Figure 14D). In contrast, cotreatment with JQ1 did not promote DMF-dependent glutathione depletion in ABC-DLBCL, possibly due to the intrinsically higher glutathione levels in ABC-DLBCL compared with GCB-DLBCL (Figure 3G; supplemental Figure 14E).14 Taken together, gene expression profiling revealed the BETi-induced downregulation of ferroptosis-protective genes, such as SLC7A11, which led to a moderate but robust reduction of the intracellular glutathione pool in GCB-DLBCL.
BRD4 controls FSP1 expression in GCB-DLBCL
Gene expression profiling of JQ1-treated DOHH2 cells revealed a reduction in AIFM2 transcript levels in response to BET inhibition (Figure 3A-B). Accordingly, treatment with other BETis, such as I-BET151, I-BET762, and PFI-1, also impaired AIFM2 mRNA in DOHH2 cells (supplemental Figure 15A). Similarly, JQ1 treatment dose-dependently decreased AIFM2 transcript levels in all other GCB-DLBCL and ABC-DLBCL cell lines investigated (Figure 4A-B; supplemental Figure 15B). As AIFM2 encodes FSP1, which plays a central role in the detoxification of lipid peroxyl radicals, we assessed FSP1 protein levels upon BET inhibition in various DLBCL cell lines by immunoblotting.21 Indeed, FSP1 protein expression was markedly impaired after the treatment with JQ1 or I-BET151, whereas GPX4 levels were unaffected or even slightly increased (Figure 4C; supplemental Figure 15C-D). Interestingly, knockdown of BRD4 using 3 independent shRNAs also resulted in reduced FSP1 protein levels, indicating that BRD4 plays a crucial role in the regulation of FSP1 expression (Figure 4D). Chromatin immunoprecipitations in SU-DHL-6 cells revealed a direct recruitment of BRD4 to the promoters of both AIFM2 and SLC7A11 (Figure 4E). To investigate whether the capacity of BRD4 to promote transcription via the P-TEFb complex is involved in the control of FSP1 expression, we treated GCB-DLBCL cells with a CDK9 inhibitor and analyzed FSP1 expression on both transcript and protein level (Figure 4F; supplemental Figure 15E-F). The observed reduction in response to CDK9 inhibition suggests that not only the function of the BRD4 bromodomains but also the recruitment and activity of the P-TEFb complex are essential to drive FSP1 expression in DLBCL.
BRD4 positively regulates FSP1 expression in GCB-DLBCL. (A-B) Various GCB-DLBCL cell lines were treated with 0.25 μM (A) or the indicated concentrations (B) of JQ1 for 24 hours. AIFM2 transcript levels were assessed by qPCR. SDHA served as reference gene. (C) FSP1 and GPX4 protein expression was analyzed by immunoblotting in various GCB-DLBCL cell lines treated with solvent, JQ1 or I-BET151 for 24 hours. GAPDH served as loading control. (D) In SU-DHL-6 and OCI-Ly7 cells, BRD4 expression was silenced using 3 independent shRNAs. FSP1 and GPX4 protein levels were visualized by immunoblotting. GAPDH served as loading control. (E) Chromatin immunoprecipitation analysis of BRD4 binding to AIFM2 and SLC7A11 promoter regions. For each gene, 2 independent promoter-specific (#1 and #2) and 1 upstream (upstr.) primer as internal negative control are shown. (F) The indicated GCB-DLBCL cell lines were treated with the CDK9 inhibitor LDC067 for 24 hours and analyzed for FSP1 expression by immunoblotting. GAPDH served as loading control. Error bars correspond to the mean ± SD. Data are representative of at least 3 (A-C) or 2 (D-F) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
BRD4 positively regulates FSP1 expression in GCB-DLBCL. (A-B) Various GCB-DLBCL cell lines were treated with 0.25 μM (A) or the indicated concentrations (B) of JQ1 for 24 hours. AIFM2 transcript levels were assessed by qPCR. SDHA served as reference gene. (C) FSP1 and GPX4 protein expression was analyzed by immunoblotting in various GCB-DLBCL cell lines treated with solvent, JQ1 or I-BET151 for 24 hours. GAPDH served as loading control. (D) In SU-DHL-6 and OCI-Ly7 cells, BRD4 expression was silenced using 3 independent shRNAs. FSP1 and GPX4 protein levels were visualized by immunoblotting. GAPDH served as loading control. (E) Chromatin immunoprecipitation analysis of BRD4 binding to AIFM2 and SLC7A11 promoter regions. For each gene, 2 independent promoter-specific (#1 and #2) and 1 upstream (upstr.) primer as internal negative control are shown. (F) The indicated GCB-DLBCL cell lines were treated with the CDK9 inhibitor LDC067 for 24 hours and analyzed for FSP1 expression by immunoblotting. GAPDH served as loading control. Error bars correspond to the mean ± SD. Data are representative of at least 3 (A-C) or 2 (D-F) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
BETi-dependent ferroptosis sensitization of DLBCL cells is mainly due to the reduction of FSP1 expression
To assess whether the downregulation of FSP1 represents the main factor responsible for the observed ferroptosis-sensitizing effect of the BETis, we tested whether treatment with a pharmacological FSP1 inhibitor (iFSP1) can mimic BRD4 inhibition and cooperate with DMF treatment in the induction of ferroptosis. Although single treatment with low doses of DMF or iFSP1 only moderately triggered lipid peroxidation in GCB-DLBCL cell lines, the combination treatment led to a strong accumulation of lipid peroxides (Figure 5A; supplemental Figure 16A). Accordingly, FSP1 silencing using 2 independent shRNAs promoted DMF-induced lipid peroxidation (Figure 5B; supplemental Figure 16B-C). In addition, the combination of DMF and iFSP1 treatment synergized to potently kill GCB-DLBCL cells (Figure 5C; supplemental Figure 17A). To further evaluate the importance of FSP1 downregulation for the ferroptosis-sensitizing function of BETis, we exogenously expressed a FLAG-tagged version of FSP1 in DOHH2 cells (supplemental Figure 17B). The resulting increase in FSP1 protein levels efficiently suppressed lipid peroxidation induced by DMF and JQ1 combinatorial treatment (Figure 5D; supplemental Figure 17C). Although JQ1 treatment sensitized cells transduced with an empty vector to DMF-induced toxicity, the synergy of the drug combination was abrogated in FSP1-overexpressing cells (Figure 5E; supplemental Figure 17D). Next, we speculated that the observed effectiveness of the combination of BETis and FINs, such as DMF and RSL3, relies on the simultaneous targeting of 2 central ferroptosis-suppressing proteins that work in parallel to restrict the accumulation of toxic lipid peroxides. While DMF depletes glutathione and thus impairs the function of GXP4, BETis decrease the expression of FSP1. To test this hypothesis, we combined iFSP1 and BETis, both targeting the activity of FSP1. Although BETis synergized with DMF and RSL3, which both inhibit GPX4 activity (Figure 2A,D), cotreatment with JQ1 did not increase the cytotoxicity of iFSP1 (Figure 5F). Collectively, these findings suggest that BETis sensitize GCB-DLBCL cells to ferroptosis mainly by downregulation of FSP1 expression and that the combined blockade of the FSP1 and GPX4 axes represents a promising strategy for antilymphoma treatment.
BET inhibitor-mediated reduction of FSP1 expression promotes ferroptosis. (A) The indicated GCB-DLBCL cell lines were treated with solvent, iFSP1, DMF, or with a combination of the compounds for 2 hours. Lipid peroxidation was quantified by flow cytometry. (B) DMF-induced lipid peroxidation was analyzed in control or AIFM2-silenced DOHH2 cells. (C) SU-DHL-6 cells were treated with DMF alone (left panel) or in combination with iFSP1 (right panel), as indicated. After 24 hours, cell numbers were determined, and the combination treatments were normalized to the DMF single treatment. The combination index (CI) for treatment with 7.5 μM DMF and 2.5 μM iFSP1 was 0.38. (D) Lipid peroxidation was quantified by flow cytometry in control or FSP1-FLAG overexpressing DOHH2 cells after combination treatment with JQ1 and DMF. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (E) DOHH2 cells were transduced with a control or an FSP1-FLAG encoding plasmid. Cell numbers upon incubation with DMF (left panels) or DMF and JQ1 (right panels) were determined and the combinatorial treatments were normalized to the DMF single treatment. (F) DOHH2 and WSU-DLCL-2 cells were treated with iFSP1 alone or in combination with the indicated JQ1 concentrations. Cell survival was assessed after 48 hours. Error bars correspond to the mean ± SD. Data are representative of at least 3 (A,C-E) or 2 (B,F) independent experiments. ∗∗P < .01; ∗∗∗P < .005. MFI, mean fluorescence intensity.
BET inhibitor-mediated reduction of FSP1 expression promotes ferroptosis. (A) The indicated GCB-DLBCL cell lines were treated with solvent, iFSP1, DMF, or with a combination of the compounds for 2 hours. Lipid peroxidation was quantified by flow cytometry. (B) DMF-induced lipid peroxidation was analyzed in control or AIFM2-silenced DOHH2 cells. (C) SU-DHL-6 cells were treated with DMF alone (left panel) or in combination with iFSP1 (right panel), as indicated. After 24 hours, cell numbers were determined, and the combination treatments were normalized to the DMF single treatment. The combination index (CI) for treatment with 7.5 μM DMF and 2.5 μM iFSP1 was 0.38. (D) Lipid peroxidation was quantified by flow cytometry in control or FSP1-FLAG overexpressing DOHH2 cells after combination treatment with JQ1 and DMF. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (E) DOHH2 cells were transduced with a control or an FSP1-FLAG encoding plasmid. Cell numbers upon incubation with DMF (left panels) or DMF and JQ1 (right panels) were determined and the combinatorial treatments were normalized to the DMF single treatment. (F) DOHH2 and WSU-DLCL-2 cells were treated with iFSP1 alone or in combination with the indicated JQ1 concentrations. Cell survival was assessed after 48 hours. Error bars correspond to the mean ± SD. Data are representative of at least 3 (A,C-E) or 2 (B,F) independent experiments. ∗∗P < .01; ∗∗∗P < .005. MFI, mean fluorescence intensity.
Combinatorial treatment with DMF and I-BET151 is effective in vivo
To test the antilymphoma efficacy of the combination of BETis and DMF in vivo, we assessed the impact of the combinatorial treatment on tumor growth in an established PDX mouse model. The VFN-D19 model was derived from a stage 4B refractory DLBCL after several (immuno-) chemotherapies (ie, R-CHOP, R-ESHAP (etoposide, Solu-Medrone [methylprednisolone], high dose Ara-C, platinol), R-ICE (ifosfamide, carboplatin, etoposide), GemOx (gemcitabine, oxaliplatin), and pixantrone/bendamustine) and was classified as GCB-DLBCL according to the Hans algorithm.34 Although DMF and I-BET151 monotherapy did not substantially interfere with tumor growth, the combined treatment with both agents significantly reduced the tumor volume (P = .008 for the combination treatment at day 10; Figure 6A). Especially during the treatment period (up to day 5) with daily application of DMF and I-BET151, the combination treatment arrested tumor growth (supplemental Figure 18A). To assess whether the drug combination induced ferroptosis in the tumor cells, we took advantage of the UPF-30P cell line, which had been established from the same patient’s primary lymphoma cells as the VFN-D19 PDX model. Although DMF treatment alone only moderately induced lipid peroxidation in UPF-30P cells, cotreatment with BETis substantially promoted the accumulation of lipid peroxides (Figure 6B). Accordingly, DMF and I-BET151 synergized in killing UPF-30P cells, which was counteracted by cotreatment with the ferroptosis suppressors ferrostatin-1 and α-tocopherol, but not with a caspase-inhibitor (Figure 6C-D). Similar to other GCB-DLBCL cell lines, the decrease in AIFM2/FSP1 expression observed in UPF-30P cells after BETi treatment presumably accounted for the ferroptosis-sensitizing effect of BET inhibition in this DLBCL model (Figure 6E-F). In agreement with this hypothesis, FSP1 inhibition promoted DMF-induced lipid peroxidation and cellular toxicity to an extent similar to BET inhibition (Figure 6G-H). Because I-BET151 treatment reduced AIFM2 transcript levels in tumors derived from VFN-D19 xenograft mice, we postulated that BETis sensitized lymphoma cells to ferroptosis also in an in vivo setting (Figure 6I). Accordingly, we detected increased levels of malondialdehyde, a biomarker for lipid peroxidation, in the tumors from mice treated with I-BET151 and DMF (Figure 6J).
BET inhibition synergizes with DMF in killing GCB-DLBCL cells in vivo. (A) VFN-D19 patient-derived xenograft mice were treated with either vehicle, DMF, I-BET151 or the combination of DMF and I-BET151 for 5 consecutive days (days 1-5), as indicated. Tumor volume was quantified by caliper measurements up to 10 days after start of the treatment. Each group consists of ≥7 animals. (B) Lipid peroxidation in UPF-30P cells treated with DMF, JQ1, I-BET151, or the indicated combinations was assessed by flow cytometry. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (C) UPF-30P cells were treated with DMF alone (left panel) or in combination with I-BET151 (right panel). After 48 hours, cell numbers were determined and the combination treatment was normalized to the DMF single treatment. The CI for treatment with 10 μM DMF and 0.25 μM I-BET151 is 0.32. (D) The potential of 10 μM ferrostatin-1 (Fer-1), 100 μM α-tocopherol (α-Toco) and 10 μM Q-VD-OPh to prevent the toxicity induced by cotreatment with DMF and I-BET151 was analyzed in UPF-30P cells. (E) Transcript levels of the indicated genes in JQ1- or I-BET151-treated UPF-30P cells were quantified by qPCR. SDHA served as reference gene. (F) FSP1 protein expression in UPF-30P cells 24 hours after JQ1 or I-BET151 treatment was visualized by immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase served as loading control. (G) UPF-30P cells were treated with solvent, DMF, and iFSP1 for 2 hours, as indicated. Lipid peroxidation was quantified by flow cytometry. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (H) Survival of UPF-30P cells after 24 hours of combinatorial treatment with DMF and iFSP1 (right panel) was normalized to the DMF single treatment (left panel). The CI for treatment with 10 μM DMF and 2.5 μM iFSP1 is 0.46. (I-J) VFN-D19 patient-derived xenograft mice were treated with either vehicle, DMF, I-BET151 or the combination of DMF and I-BET151 (n = 3 for each group) for 3 consecutive days. Subsequently, the respective tumors were isolated und analyzed for AIFM2 mRNA expression by qPCR (I) or malondialdehyde levels by thiobarbituric acid reactive substances (TBARS) assay (J). Error bars correspond to the mean ± SD. Data are representative of at least 3 (B-F,H) or 2 (G) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. CI, combination index; MFI, mean fluorescence intensity; qPCR, quantitative polymerase chain reaction.
BET inhibition synergizes with DMF in killing GCB-DLBCL cells in vivo. (A) VFN-D19 patient-derived xenograft mice were treated with either vehicle, DMF, I-BET151 or the combination of DMF and I-BET151 for 5 consecutive days (days 1-5), as indicated. Tumor volume was quantified by caliper measurements up to 10 days after start of the treatment. Each group consists of ≥7 animals. (B) Lipid peroxidation in UPF-30P cells treated with DMF, JQ1, I-BET151, or the indicated combinations was assessed by flow cytometry. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (C) UPF-30P cells were treated with DMF alone (left panel) or in combination with I-BET151 (right panel). After 48 hours, cell numbers were determined and the combination treatment was normalized to the DMF single treatment. The CI for treatment with 10 μM DMF and 0.25 μM I-BET151 is 0.32. (D) The potential of 10 μM ferrostatin-1 (Fer-1), 100 μM α-tocopherol (α-Toco) and 10 μM Q-VD-OPh to prevent the toxicity induced by cotreatment with DMF and I-BET151 was analyzed in UPF-30P cells. (E) Transcript levels of the indicated genes in JQ1- or I-BET151-treated UPF-30P cells were quantified by qPCR. SDHA served as reference gene. (F) FSP1 protein expression in UPF-30P cells 24 hours after JQ1 or I-BET151 treatment was visualized by immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase served as loading control. (G) UPF-30P cells were treated with solvent, DMF, and iFSP1 for 2 hours, as indicated. Lipid peroxidation was quantified by flow cytometry. The MFI of oxidized BODIPY C11 in treated cells was normalized to the MFI of the solvent control. (H) Survival of UPF-30P cells after 24 hours of combinatorial treatment with DMF and iFSP1 (right panel) was normalized to the DMF single treatment (left panel). The CI for treatment with 10 μM DMF and 2.5 μM iFSP1 is 0.46. (I-J) VFN-D19 patient-derived xenograft mice were treated with either vehicle, DMF, I-BET151 or the combination of DMF and I-BET151 (n = 3 for each group) for 3 consecutive days. Subsequently, the respective tumors were isolated und analyzed for AIFM2 mRNA expression by qPCR (I) or malondialdehyde levels by thiobarbituric acid reactive substances (TBARS) assay (J). Error bars correspond to the mean ± SD. Data are representative of at least 3 (B-F,H) or 2 (G) independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005. CI, combination index; MFI, mean fluorescence intensity; qPCR, quantitative polymerase chain reaction.
Collectively, we demonstrate that in GCB-DLBCL cells, BETi impairs the expression of FSP1 and thus sensitizing the lymphoma cells to ferroptosis. Our data suggest that the combination of BETis and GPX4-targeting FINs could represent a promising novel strategy in DLBCL therapy.
Discussion
In recent years, there has been a growing interest in harnessing ferroptosis induction to circumvent apoptosis resistance and treat tumors refractory to conventional therapy approaches.17,35 The treatment of a panel of 117 cancer cell lines with the SLC7A11 inhibitor erastin identified DLBCL as one of the tumor entities that exhibits the greatest vulnerability to ferroptosis.15 The molecular basis for this profound ferroptosis sensitivity is, however, not well-understood and might involve several cellular characteristics. For instance, it has been reported that the trans-sulfuration pathway, which generates cysteine from homocysteine or methionine, is often defective in lymphoma.36-38 In line, Schmitt et al reported that the low intracellular glutathione levels observed especially in GCB-DLBCL cell lines were easily depleted by treatment with the electrophile DMF, leading to impaired GPX4 activity, massive lipid peroxidation, and induction of ferroptosis.14 Moreover, GCB-DLBCL cells exhibit high expression of arachidonate 5-lipoxygenase that is capable of enzymatically catalyzing lipid peroxidation and potentially contributes to the increased ferroptosis susceptibility of this DLBCL subtype.14
Although FINs, such as DMF or imidazole ketone erastin, have been shown to impair the growth of lymphoma cells in preclinical in vivo models, they often lack efficacy to arrest tumor growth or to even eradicate the tumor.14,39 Thus, we aimed to identify chemical compounds that can boost the effect of the FINs and were able to demonstrate that BRD4 inhibition potently sensitizes DLBCL cells to ferroptosis. As BET proteins drive the expression of oncogenes, such as MYC, and MYC amplification is a common event in hematological malignancies, BETis have been tested in clinical trials for the treatment of lymphoma, leukemia, and myeloma.26,29,40,41 However, the monotherapy with BETis exhibited only a limited efficacy which did not correlate with MYC expression.41 This discrepancy might be explained by the fact that several previous preclinical studies have used clinically irrelevant high doses of the BETis, which cause a general alteration of polymerase II–dependent transcription and thus, might contribute to the misinterpretation of their anticancer potential.41,42
In contrast to previous studies that had demonstrated a JQ1-dependent downregulation of GPX4 or the induction of ferritinophagy in breast cancer cell lines, we were unable to detect these effects in DLBCL in response to BET inhibition (Figure 3B-C; supplemental Figure 18B).43,44 Whether cell type–specific characteristics or different BETi concentrations are responsible for these disparities, should be addressed in future studies. Here, we demonstrate that inactivation of BRD4 by shRNA-mediated silencing or treatment with BETis in the nanomolar range reduced the expression of ferroptosis-associated proteins, such as FSP1 and SLC7A11, in DLBCL cells. Strikingly, the impaired expression of FSP1 and SLC7A11 after BRD4 inhibition was not sufficient to provoke lipid peroxidation on its own, but rather sensitized the cells to the treatment with GPX4-targeting FINs. As BETi-dependent downregulation of SLC7A11 only partially reduced the cellular glutathione pool in DLBCL, we propose that the observed decrease in FSP1 expression constitutes the major cause of the ferroptosis sensitization induced by BET inhibition. This is in line with our observation that pharmacological inhibition or knockdown of FSP1 mimicked the effects of BETis and similarly increased ferroptosis susceptibility, whereas expression of exogenous FSP1 abrogated the synergy of BETi and DMF combinatorial treatment. In contrast, the combination of inhibitors targeting FSP1 and BET proteins did not result in synergistic killing of DLBCL cells. Therefore, it is tempting to speculate that the synergy observed for the BETi and DMF combination is mainly mainly due to simultaneous targeting of 2 key enzymes that are crucial for the detoxification of lipid peroxides. While DMF inhibits GPX4 by depleting glutathione, BET inhibition reduces FSP1 expression and thus impairs the regeneration of important antioxidative entities, such as coenzyme Q10, vitamin K, and α-tocopherol, which can scavenge lipid peroxyl radicals.14,21,22 This hypothesis is supported by the observation that low FSP1 expression in cancer cells correlates with an increased GPX4 dependency and that in some tumor cells ferroptosis can only be provoked by targeting both enzymes.21,45
Recent studies have suggested that tumor cells surviving the sublethal exposure to apoptosis-inducing compounds become drug-tolerant persister cells.46 Strikingly, this persister phenotype is correlated with an increased vulnerability to ferroptosis, highlighting the therapeutic potential of ferroptosis-inducing treatments. Our data provide a rationale to clinically evaluate the combination of DMF or other GPX4-targeting FINs and BETis for the treatment of GCB-DLBCL. Although DMF is a Food and Drug administration– and European Medicines Agency–approved drug used in the treatment of psoriasis and relapsing-remitting multiple sclerosis and is well-tolerated even during long-term application, the safety and efficacy of BETis are currently evaluated in patients with hematological malignancies and solid tumors. So far, dose-limiting toxicities, such as thrombocytopenia, anemia, and gastrointestinal disorders, have been observed in patients treated with BETis.41,47-49 As low concentrations of BETis (ie, 100 nM JQ1) were sufficient to reduce AIFM2/FSP1 expression and to promote DMF-induced lipid peroxidation and toxicity (Figure 2A and Figure 4B), it might be feasible to achieve a sensitization of the tumor cells to ferroptosis with tolerable doses of the BETis. Thus, the use of BETis in combination with GPX4-targeting FINs, such as DMF, could represent a promising novel therapeutic strategy in the treatment of patients with DLBCL.
Acknowledgments
The authors thank Maik Franke, Corinna Kosnopfel, Wendan Xu, and Marcus Conrad for helpful discussion and advice. This work was supported by the DFG Collaborative Research Center Transregio SFB/TR 156 (to S.H.), the Deutsche Krebshilfe (to G.L. and S.H.), the Innovative Medizinische Forschung (SC122215) of the University of Münster (to A.S.), the ETH Research Grant funding (ETH-33 19-2, to M.Z.), the Ministry of Health of the Czech Republic (grant AZV NU21-03-00386), and the Grant Agency of the Czech Republic (grant GA23-05474S) (both to P.K.).
Authorship
Contribution: A.S., M. Grimm, N.K., H.J., J.L., L.S., C.S., E.G., A.T., K.M. and A.D. performed experiments; A.S., A.B., C.L., F.R., M. Grau, M.Z., K.S.-O., P.K., G.L. and S.H. analyzed data; A.S., G.L. and S.H. wrote the manuscript, which all other authors commented on; and A.S., G.L., and S.H. conceived and coordinated the study.
Conflict-of-interest disclosure: G.L. has participated in a consulting or advisory role for AbbVie, ADC Therapeutics, Amgen, AstraZeneca, Bayer, BMS, Constellation, Gilead, Genase, Genmab, Hexal/Sandoz, Immagene, Incyte, Janssen, Karyopharm, Miltenyi, Morphosys, NanoString, Novartis, PentixaPharm, Roche, Sobi and Takeda, has been on a speaker’s bureau for Bayer, Celgene, Gilead, Janssen and Roche, and has received research funding from AQUINOX, AstraZeneca, Bayer, Celgene, Gilead, Janssen and Roche. P.K. has participated in a consulting or advisory role for Eli-Lilly, Takeda and Janssen, and received research funding from AQUINOX, AstraZeneca and MorphoSys. The remaining authors declare no competing financial interests.
Correspondence: Stephan Hailfinger, Department of Medicine A, Translational Oncology, University Hospital Münster, Domagkstr. 3, 48149, Münster, Germany; e-mail: stephan.hailfinger@ukmuenster.de.
References
Author notes
Gene expression and mass spectrometry data are available at Gene Expression Omnibus (accession number GSE231871) and the European Molecular Biology Laboratory - European Bioinformatics Institute database (EMBL-EBI, S-BSST1072), respectively.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal