Abstract
In this spotlight, we review technical issues that compromise single-cell analysis of tissue macrophages, including limited and unrepresentative yields, fragmentation and generation of remnants, and activation during tissue disaggregation. These issues may lead to a misleading definition of subpopulations of macrophages and the expression of macrophage-specific transcripts by unrelated cells. Recognition of the technical limitations of single-cell approaches is required in order to map the full spectrum of tissue-resident macrophage heterogeneity and assess its biological significance.
Introduction
Mononuclear phagocytes are a family of cells comprising progenitors, blood monocytes, dendritic cells (DCs), and resident tissue macrophages. The proliferation, differentiation, and survival of mononuclear phagocytic cells is controlled by signals from the macrophage colony-stimulating factor receptor (CSF1R).1 Localization using a knockin transgene in the mouse Csf1r locus highlights their surprisingly uniform density in every tissue.2 Tissue macrophages share many features, including cell shape, association with basement membranes, and phagocytic function. In individual tissues, and specific niches within certain tissues macrophage gene expression adapts to support specific functions.3,4 Macrophage heterogeneity within and between mouse tissues was previously defined through the use of monoclonal antibodies against cell-surface markers.5 More recently, transgenic mouse lines directly expressing fluorescent reporters or Cre recombinase in combination with conditional reporters have been based upon an ever-expanding array of myeloid-enriched genes (eg, Adgre1, Ccr2, Cd68, Clec9a, Crybb1, Csf1r, Cx3cr1, Cxcr4, Flt3, HexB, Irf8, Itgam, Itgax, Kit, LysM, Ly75, Ms4a3, Mafb, Mrc1, Runx1, Slco2b1, Spi1, Sall1, S100a4, Siglec1, Tnfrsf11a (RANK), and Zbtb46). These tools enable visualization of mouse mononuclear phagocyte subpopulations in situ, including the still-contentious distinction between macrophages and DCs in nonlymphoid tissues.3,6,7 Myeloid-specific tamoxifen-inducible Cre recombinase transgenes have been used in fate-mapping to dissect the relative importance of self-renewal vs replacement by monocytes in tissue-resident macrophage homeostasis (as previously reviewed8). Some assumptions underlying fate-mapping in inbred mice have been questioned, suggesting that the contribution of monocytes may be underestimated.7 Regardless, monocytes are clearly able to replace resident tissue macrophages when the tissue niche is vacant.7,9 The advent of single-cell technologies applied to mononuclear phagocytes isolated by tissue disaggregation has led to the subdivision of resident and recruited macrophages and DCs in the mouse into subpopulations.10-13 This brief review identifies key caveats that compromise the interpretation of single-cell data and strategies to mitigate potential artifacts.
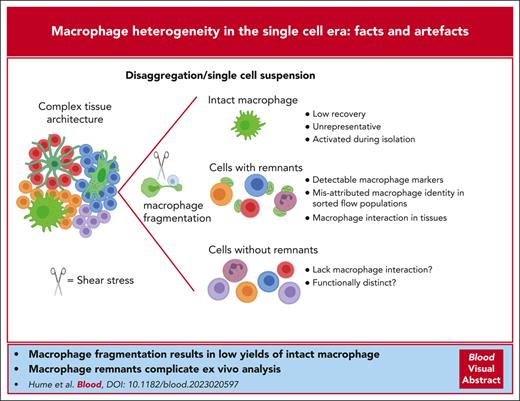
Does tissue disaggregation produce a representative sample of tissue macrophages?
Single-cell RNA-seq (scRNA-seq) analysis relies upon tissue disaggregation to generate single-cell suspensions. Macrophages are so abundant in every tissue that their gene expression profile can be detected in total tissue RNA-seq data. Indeed, the signatures of resident macrophages can be extracted by comparing total RNA-seq data from wild-type and Csf1r knockout mice and rats.14,15 Based upon the relative expression of macrophage-specific genes, such as Csf1r, in purified macrophages and tissues, we estimate that macrophages contribute 5% to 15% of the total messenger RNA (mRNA) in every tissue, consistent with the cellular abundance detected with Csf1r reporter transgenes.2 Against that background, the yield of macrophages after tissue disaggregation in mice is much less than 10% of the numbers in situ.16,17 Well-defined resident populations, such as those of the bone and bone marrow, marginal zone of the spleen, and sinuses of lymph nodes, have never been isolated intact.17,18 The definitive marker of these cells in situ, Siglec1 (or CD169)19,20 is not detected in major tissue scRNA-seq resources, including the Tabula Muris21 and Mouse Cell Atlas.22 In these resources, few cells are annotated as macrophages, even in macrophage-rich tissues, such as the spleen. Similarly, macrophage clusters represent <2% of cells in hematopoietic tissues in the human cell atlas.23 Those clusters lack detectable expression of markers such as MERTK, TIMD4, SIGLEC1, VCAM1, CX3CR1, ITGAM, and CD163, and only a subset has detectable CSF1R. In overview, disaggregation procedures fail to release representative populations of resident macrophages.
Does tissue disaggregation permit the isolation of intact macrophages?
The extensive arborization of membrane processes in tissue macrophages2 and their adhesion to extracellular matrix and other cells likely ensure that even the gentlest tissue disaggregation procedures fragment the cells. Fragmentation was first described in subcapsular sinus macrophages with membrane bleb attachment to IL17R-expressing lymphocytes.24 Remarkably, disrupted macrophage fragments can reseal and retain mRNA and protein, including surface markers, to generate remnants attached to other cells in a pattern that reflects their juxtaposition in situ.17 The incidence of macrophage remnants attached to unrelated cells in the spleen and marrow was reduced in CD169-deficient mice, indicating the importance of cell-cell interaction.17 Many cells that appear in the macrophage gate on a typical flow cytometry profile of hematopoietic tissues are unrelated cells with 1 or more attached macrophage remnants.17 The impacts of macrophage fragmentation are evident in published macrophage transcriptomes. Meta-analysis of total RNA-seq data from macrophages isolated from all major organs revealed high levels of contamination with mRNA from unrelated cells.3 Some of the contamination may also derive from their active roles in the clearance of apoptotic or senescent cells. For example, most isolated macrophages have detectable hemoglobin mRNA reflecting erythrophagocytosis, and epithelial markers in gut lamina propria macrophages may reflect a role in homeostatic turnover.3
In contrast, unrelated cells may appear to express macrophage-related transcripts. In both Tabula Muris and Mouse Cell Atlas, 20% to 30% of neurons, astrocytes, and oligodendrocytes apparently express macrophage/microglia-specific genes, including Csf1r. Given the clear evidence that Csf1r is expressed only in mononuclear phagocytes2 and the selective depletion of the microglial signature in Csf1r mutant mice,15 this is most likely an artifact.
Macrophages and remnants in the bone marrow
Fragmentation of resident macrophages in bone marrow has compromised studies of their roles in the regulation of hematopoiesis.17 Baccin et al25 intended to generate a molecular map of all major bone marrow cell populations to inform bone marrow niche organization, including strategies to enrich for rare cell types. Illustrating the limitations of tissue disaggregation, resident macrophages, indisputably a major cell population in the marrow that contributes to the development of specialized bone marrow niches as well as bone mineral homeostasis,17 are absent from their analyses and commentary.25 The isolation and characterization of erythroblastic island (EBI) macrophages demonstrates the impacts of fragmentation. EBI macrophages were previously distinguished from other bone marrow macrophages based on coexpression of F4/80 and the granulocyte marker Ly6G.17 Two recent studies in Blood26,27 described EBI macrophages from mouse bone marrow. In both cases, there was evidence of poor yield of resident macrophages (eg, the absence of detectable Siglec1 mRNA encoding the CD169 marker17). Reanalysis of the data from Li et al26 indicated extensive contamination of the purified macrophages with granulocyte-specific transcripts.3 Romano et al27 applied cellular indexing of transcriptomes and epitopes and scRNA-seq analysis to EBI clusters and detected 28 transcriptionally distinct populations, including multiple clusters identified as lymphocytes or mast cells. Despite the purification of the EBI before dispersal, intact F4/80+ macrophages remained a minor subpopulation, apparently expressing gene signatures of other hematopoietic lineages. Only a subset expressed a transcriptional profile consistent with their own27 and previously published28 EBI imaging data. With imaging flow cytometry, F4/80+Ly6G+ bone marrow events are without exception Ly6G+ granulocyte lineage cells with attached macrophage remnants,17 consistent with the conclusion that EBI macrophages also support granulopoiesis.27 EBI macrophages have also been identified as a minor population in the context of scRNA-seq analysis of hematopoiesis in human fetal liver.29 However, the accompanying flow cytometry data suggest that many of these cells are either doublets or remnants associated with erythroid cells.
Marrow macrophage fragmentation also contributes to the ambiguity surrounding markers expressed by hematopoietic stem cells (HSCs) and committed progenitors. A recent study identified VCAM1 on isolated HSC,30 extending earlier claims of the existence of subsets of HSCs that express mature macrophage markers CSF1R and F4/80.31,32 In other studies, Csf1r mRNA and an integrated Csf1r reporter transgene were not detected in HSCs. They were first detected in a small portion of hematopoietic progenitor cells and increased progressively in committed progenitors.2,17,33 We found that the F4/80 antigen detected in HSCs derives entirely from attached macrophage remnants.17Figure 1A-C shows imaging flow cytometry demonstrating that VCAM1 detection in HSC events in mouse marrow is also macrophage remnant–derived, confounding the suggestion that VCAM1 confers immune tolerance on transplanted cells.30 A related concept is the recent detection of major histocompatibility complex class II (MHCII) expression on HSCs based upon global transcriptome datasets, flow cytometry, quantitative reverse transcription polymerase chain reaction on sorted cells, ex vivo assessment of freshly isolated cells, and a proposed antigen-presentation capacity.34Figure 1D-E identifies a subset of HSCs with cell-surface MHCII staining consistent with the expression of H2-Aa and Cd74 detected in HSCs that lack macrophage markers.33 However, most MHCII membrane staining appears as remnant derived. Most cell isolation procedures for hematopoietic tissues use simple mechanical disaggregation, but enzyme digestion does not reduce the incidence of remnants. Figure 1F shows the profile of cells isolated by enzymatic digestion of bone marrow plugs. The analysis of bone marrow highlights both the analytical challenge that macrophage remnants present for cellular phenotyping and the opportunity to deconvolute scRNA-seq data to reveal cell-cell interactions.
VCAM1 and MHCII staining on hematopoietic stem and progenitor cell (HSPC) populations is predominantly remnant associated. Imaging flow cytometry analysis was performed on HSPC populations in Kit-enriched bone marrow (BM) from C57BL/6J mice as described,17 except that the anti-F4/80 antibody was replaced with the anti-VCAM1 or anti-MHCII antibody, as indicated. (A) VCAM1 was detected on most cells in all HSPC subsets: Lin–Sca1+Kit+CD48–CD150+ HSCs, Lin–Sca1+Kit+CD48–CD150– multipotent progenitors (MPPs), Lin–Sca1+Kit+CD48+ hematopoietic progenitor cells (HPCs), and Lin–Sca1–Kit+ committed progenitors (CPs). (B) Representative images of HSCs display cell-surface staining of population-defining markers (Sca1, Kit, and CD150), contrasting with punctate, remnant-associated VCAM1 staining. Violin plots of stain area for each marker (n = 600 cells) demonstrate that VCAM1 stain area was consistently lower than that of Sca1, Kit, and CD150. (C) Manual classification reveals the VCAM1 staining pattern on HSCs, MPPs, and HPCs to be exclusively remnant associated (>300 cells classified per subset), whereas a small fraction of VCAM1+ CPs display cell-surface VCAM1 expression. For each classification, representative images of VCAM1+ CP are displayed as a single-color fluorescent image and a brightfield overlay with the same image. (D-F) (D) MHCII were detected on most HSCs, MPPs, and HSCs and at a lower frequency on CPs. (E) Manual classification of MHCII staining patterns on HSPC subsets (>500 cells classified/subset), with representative images of MHCII+ HSC for each classification. (F) Imaging flow cytometry of BM single-cell suspension generated by collagenase type IV/DNase I digestion (37°C for 40 minutes) of an extruded tibial BM plug, followed by gentle mechanical disruption with a pipette, and assessed for CD11b, F4/80, and VCAM1 expression. (Left) Scatter plot displaying F4/80 and VCAM1 staining intensity for all focused singlets. The pink gate indicates double-positive events, and cells manually classified as having surface staining for both F4/80 and VCAM1 (less than 3%) are highlighted in yellow on the scatter plot. The middle panel shows representative images for abundant remnant F4/80+VCAM1+ staining patterns, with the left panel showing represented images of the infrequent cells exhibiting surface F4/80+VCAM1+ staining pattern. These data are representative of the outcome of ex vivo digestion strategies tested on both the BM and spleen, with no substantive gains made in intact macrophage recovery from these hematopoietic tissues. Scale bar, 10 μm. BF, Bright Field.
VCAM1 and MHCII staining on hematopoietic stem and progenitor cell (HSPC) populations is predominantly remnant associated. Imaging flow cytometry analysis was performed on HSPC populations in Kit-enriched bone marrow (BM) from C57BL/6J mice as described,17 except that the anti-F4/80 antibody was replaced with the anti-VCAM1 or anti-MHCII antibody, as indicated. (A) VCAM1 was detected on most cells in all HSPC subsets: Lin–Sca1+Kit+CD48–CD150+ HSCs, Lin–Sca1+Kit+CD48–CD150– multipotent progenitors (MPPs), Lin–Sca1+Kit+CD48+ hematopoietic progenitor cells (HPCs), and Lin–Sca1–Kit+ committed progenitors (CPs). (B) Representative images of HSCs display cell-surface staining of population-defining markers (Sca1, Kit, and CD150), contrasting with punctate, remnant-associated VCAM1 staining. Violin plots of stain area for each marker (n = 600 cells) demonstrate that VCAM1 stain area was consistently lower than that of Sca1, Kit, and CD150. (C) Manual classification reveals the VCAM1 staining pattern on HSCs, MPPs, and HPCs to be exclusively remnant associated (>300 cells classified per subset), whereas a small fraction of VCAM1+ CPs display cell-surface VCAM1 expression. For each classification, representative images of VCAM1+ CP are displayed as a single-color fluorescent image and a brightfield overlay with the same image. (D-F) (D) MHCII were detected on most HSCs, MPPs, and HSCs and at a lower frequency on CPs. (E) Manual classification of MHCII staining patterns on HSPC subsets (>500 cells classified/subset), with representative images of MHCII+ HSC for each classification. (F) Imaging flow cytometry of BM single-cell suspension generated by collagenase type IV/DNase I digestion (37°C for 40 minutes) of an extruded tibial BM plug, followed by gentle mechanical disruption with a pipette, and assessed for CD11b, F4/80, and VCAM1 expression. (Left) Scatter plot displaying F4/80 and VCAM1 staining intensity for all focused singlets. The pink gate indicates double-positive events, and cells manually classified as having surface staining for both F4/80 and VCAM1 (less than 3%) are highlighted in yellow on the scatter plot. The middle panel shows representative images for abundant remnant F4/80+VCAM1+ staining patterns, with the left panel showing represented images of the infrequent cells exhibiting surface F4/80+VCAM1+ staining pattern. These data are representative of the outcome of ex vivo digestion strategies tested on both the BM and spleen, with no substantive gains made in intact macrophage recovery from these hematopoietic tissues. Scale bar, 10 μm. BF, Bright Field.
Macrophage remnants in other locations
Macrophage fragmentation affects the characterization of macrophages in other tissues. Lynch et al35 reported that negative selection to remove sinusoidal endothelial cells (LSECs) was a necessary step in the purification of Kupffer cells (KCs), the resident macrophages of the liver. Three recent papers36-38 identified a Kupffer cell subset (KC2) that appeared to coexpress KC-specific transcripts and markers of LSECs. In response to criticism of the original studies,39 the authors highlighted the detection of subsets of KC using an Mrc1-cre conditional reporter40 and argued against the idea that KC2 are KCs with attached LSEC remnants. The alternative proposal is that KC2 are, in fact, LSECs with attached KC remnants,39,41 reflecting the direct and intimate interaction between the 2 cell types in situ.42 This proposal was supported by 3-dimensional reconstruction, which revealed unequivocal separation of the LSEC marker GR182 and the KC marker F4/80.39 LSEC markers were not detected in the total RNA-seq profiles of isolated KCs from 2 independent groups.42,43 scRNA-seq analysis of liver cell populations by another laboratory44 concluded that proposed markers of KC2 (Esam, Clec4g, Gpr182, Stab2, and Ptprb) are LSEC restricted, whereas Mrc1 (used as a differential KC2 marker) was expressed equally and uniformly by both LSECs and KCs. Another major liver scRNA-seq data set removed as doublets, cells in which both LSEC (eg, Gpr182, Clec4g, Ptprb, and Kdr) and macrophage markers (eg, Adgre1, Clec4f, and Timd4) were detected.45 The nonoverlapping expression of KC and LSEC markers is evident in both the Tabula Muris and Mouse Cell Atlas and in analysis of a multiparameter separation pipeline, in which Adgre1 (F4/80) and the endothelial marker Cyp4b1 (enriched in KC237) were mutually exclusive.46 The published studies of KC heterogeneity differ significantly in precisely how livers are digested and disaggregated and in the markers used to separate cells by flow cytometry. Regardless of the origin, the identification of apparent hybrid cells could reflect intimate interactions in vivo. KC2 could be a subset of KCs that is especially intercalated with LSECs.
Another relevant example posits the existence of mesenchymal stem cells that express myeloid markers, including Aif1 and Csf1r.47 This work builds upon an analysis of cancer-associated fibroblasts, which included the claim that primary lung fibroblasts express CSF1R.48 This is clearly not the case in vivo.2 Similarly, the recent identification of neuron-like tumor-associated macrophages should be considered with caution.49
Interstitial lung macrophages: a case study for the interpretation of scRNA-seq data
The lung contains at least 4 distinct mononuclear phagocyte populations: bronchoalveolar macrophages (AMs) and interstitial macrophages (IMs), extravasating monocytes, and cells annotated as DCs.50,51 Lung scRNA-seq data highlight issues that affect similar analysis in many other tissues. The first issue is whether isolated cells are representative of tissue populations. Regardless of gating strategy, in myeloid populations derived from lung disaggregation, IMs are a very minor subpopulation compared with AM, at most 10% of the total pulmonary macrophages. By contrast, localization of Csf1r reporter transgenes in mouse and rat highlighted the presence of an abundant ramified IM population surrounding the alveoli,2,52,53 at least as abundant as AMs. Figure 2A shows examples of whole-mount images of mouse lung, highlighting the relative abundance and morphology of IMs. The paucity of IMs after lung disaggregation is also evident in a recent integrated cell atlas of the human lung54 (Figure 2B). Analysis of human lung macrophages by quantitative stereology confirmed that IMs are in excess compared with AMs but are drastically underrecovered after tissue disaggregation.55 Taking all of the data together, we suggest that tissue disaggregation in the lung releases only the minor population of IMs associated with connective tissues surrounding the major airways, whereas, in common with CD169+ populations in hematopoietic tissues, the major interstitial population is fragmented in the process of disaggregation.
Macrophages isolated from the lung are not representative of their abundance in vivo and are activated during the isolation process. (A) Maximum-intensity projections of whole-mount confocal images of an intact adult mouse lung from Csf1r-FusionRed mice2 (left) and from Siglec1-cre x Rosa26-ZSGreen mice (right). Note the stellate morphology of abundant IMs surrounding terminal alveoli. The Csf1r transgene detects IMs, whereas the brighter CD169 transgene detects both IMs and round alveolar macrophages (arrows) with similar relative abundance. (B) The cellular annotations of human lung leukocyte populations from the integrated human lung cell atlas.54 Note the minor subpopulation annotated as interstitial Mph perivascular (IM). A separate annotation of cDC2 is based upon the expression of CD1C, CLEC10A, and FCER1A, all of which are inducible in monocytes.56 IMs were distinguished from monocyte-derived macrophages based upon the expression of F13A1 and FOLR2.54 (C) screen shots of data of the individual genes indicated from the complete human lung atlas viewed in CellXGene. Blue dots show cells with high expression of the indicated transcripts. Cellular annotations from the atlas are indicated in red. There is no defined IM population in this projection. CSF1R and MARCO appear mutually exclusive, with the latter being associated with alveolar macrophages and CSF1R detected weakly in an overlapping set of clusters annotated as monocytes, elicited macrophages, and lung macrophages. FOS (and other immediate early genes, including FOSB, JUN, and EGR1, not shown) are detected throughout the hematopoietic, endothelial, mesenchymal, and epithelial compartments. Bottom panels show representative inducible cytokines and chemokines apparently expressed by subpopulations of AMs and other macrophages (IL1B) as well as T cells (CCL3). Arrows highlight the clusters of expressing cells.
Macrophages isolated from the lung are not representative of their abundance in vivo and are activated during the isolation process. (A) Maximum-intensity projections of whole-mount confocal images of an intact adult mouse lung from Csf1r-FusionRed mice2 (left) and from Siglec1-cre x Rosa26-ZSGreen mice (right). Note the stellate morphology of abundant IMs surrounding terminal alveoli. The Csf1r transgene detects IMs, whereas the brighter CD169 transgene detects both IMs and round alveolar macrophages (arrows) with similar relative abundance. (B) The cellular annotations of human lung leukocyte populations from the integrated human lung cell atlas.54 Note the minor subpopulation annotated as interstitial Mph perivascular (IM). A separate annotation of cDC2 is based upon the expression of CD1C, CLEC10A, and FCER1A, all of which are inducible in monocytes.56 IMs were distinguished from monocyte-derived macrophages based upon the expression of F13A1 and FOLR2.54 (C) screen shots of data of the individual genes indicated from the complete human lung atlas viewed in CellXGene. Blue dots show cells with high expression of the indicated transcripts. Cellular annotations from the atlas are indicated in red. There is no defined IM population in this projection. CSF1R and MARCO appear mutually exclusive, with the latter being associated with alveolar macrophages and CSF1R detected weakly in an overlapping set of clusters annotated as monocytes, elicited macrophages, and lung macrophages. FOS (and other immediate early genes, including FOSB, JUN, and EGR1, not shown) are detected throughout the hematopoietic, endothelial, mesenchymal, and epithelial compartments. Bottom panels show representative inducible cytokines and chemokines apparently expressed by subpopulations of AMs and other macrophages (IL1B) as well as T cells (CCL3). Arrows highlight the clusters of expressing cells.
Despite the apparent low abundance of recovered IMs, single-cell approaches have been used to define IM subpopulations based upon the expression of transcripts encoding cell-surface markers, including Mrc1 (CD206) MHCII, Lyve1, Cx3cr1, and Siglec1.12,57-61 Distinct IM subpopulations defined by markers were proposed to associate with nerves and major airways.57,58,61 Chakarov et al57 extended their finding in the lung to posit the existence of Lyve1high and MHCIIlow macrophage subsets in other tissues, including adipose, heart, and skin. A meta-analysis of macrophage total RNA-seq and scRNA-seq data from multiple tissues called into question the existence of this subset in that the proposed markers were not stringently correlated with each other.3
Macrophage activation during tissue disaggregation
The other major issue exemplified in published mouse lung data is that macrophages are activated during the process of isolation.3 The IM populations discussed earlier express very high levels of inducible immediate early transcripts (Fos, Jun, Dusp1, Egr1, and Nr4a1) and inflammatory cytokines and chemokines (Il1b, Cxcl1, and Ccl2/3/4). None of these transcripts is detected in mouse total tissue mRNA.62 The same issue is evident in human lung data. Figure 2C shows the profiles of marker genes in the human lung cell atlas.54 Inducible genes such as FOS, IL1B, and CCL3 appear more highly expressed in subsets than the macrophage-specific receptor CSF1R or AM marker MARCO but are undetectable in total human lung mRNA, in which CSF1R is relatively abundant.62 The inducible genes are so highly expressed that they occupy a significant proportion of transcriptional space in isolated macrophages3 and may compromise detection of other transcripts by scRNA-seq, leading to the designation of subsets based upon their expression. A subset of AM defined by CCL3 expression (Figure 2B) is most likely an artifact of activation during isolation. Similarly, a recent study defined 7 subpopulations of resident macrophages in the mouse kidney, separated almost entirely based upon known inducible genes63 that are not detectable in total kidney mRNA.62 In general, in the absence of detectable expression in total tissue mRNA, any macrophage population expressing immediate early genes or inflammatory cytokines has likely been activated during isolation.
Stochastic sampling vs probabilistic gene expression
Typical scRNA-seq data have a high proportion of zeros. Most analytical pipelines classify zeros as drop-outs. Variation in the detection of individual transcripts within a cell type by scRNA-seq is attributed to the Poisson distribution of detection at relatively low coverage.64 Because the relative abundance of mRNA transcripts shows a log-normal distribution, most of the transcriptomic space is occupied by the most highly expressed transcripts.3 In macrophages, that set includes transcripts encoding cell-surface receptors, presumably because the protein products have relatively high turnover rates. However, transcription is a probabilistic process that occurs in bursts at individual loci.65-67 The impact of transcription probability on variation in mRNA abundance within a cell population depends upon the average intervals between bursts of transcription and the stability of the mRNA. Extreme variation in the level of mRNA does not necessarily correlate with protein variation if the protein product is more stable than the mRNA. Transcriptomic heterogeneity has been analyzed in vitro in bone marrow cells grown in CSF2 (granulocyte macrophage CSF), a complex mixture of differentiation states mostly expressing macrophage markers.68 Shalek et al69,70 used this system and scRNA-seq to demonstrate extreme variation in transcript induction among individual cells stimulated for 4 hours with lipopolysaccharide (LPS), validated by the detection of mRNA using in situ hybridization. All-or-nothing gene expression was not restricted to inducible cytokines and chemokines and was, apparently, gene autonomous. Similar stochastic variation has even been reported in a mouse macrophage cell line response to LPS.71 The variation in mRNA abundance can potentially arise from at least 2 sources. Some genes may be sequestered in inactive chromatin and unavailable for transcription. For others, in which the mRNA is relatively unstable, variation may simply be a matter of timing since the receipt of an extracellular signal and/or the most recent transcriptional burst. In summary, many zero expression values in macrophage scRNA-seq data may be due to genuine variation that reflects the nature of transcriptional regulation.
Subpopulations vs trajectories
Subpopulations of blood monocytes, defined in mice by the expression of Ly6C, are clearly a differentiation series controlled by CSF1R signaling.6 It is less clear how subpopulations of tissue macrophages are related to each other. A dense time course of the response of human macrophages to LPS revealed multiple successive waves of enhancer activation, inducible gene expression, and feedback and feedforward activation.56 In scRNA-seq analysis, each of these intermediate states could define a subpopulation. Various computational approaches can be used to infer differentiation trajectories even from sampling of populations of cells at a single time point.72 Sanin et al11 applied trajectory analysis to diverse scRNA-seq data sets in the mouse to infer the existence of 4 conserved monocyte activation paths. They focused, in part, on the ability of recruited cells to adopt a resident macrophage phenotype. Their analysis raises the more general issue of the response of resident macrophages during inflammation. All of the transcripts that are commonly considered as markers of anti-inflammatory macrophage polarization are actually highly expressed by resident tissue macrophages,3 which may also proliferate in response to inflammatory stimuli.7,73 As noted in the introduction, resident macrophages may be replaced by self-renewal or monocyte infiltration. Regardless of the mechanism of replacement, the gene expression profile of each resident macrophage (eg, expression of Timd4) likely reflects, in part, their life history73 and is not necessarily related to any specific function.
Mitigation of artifacts and validation approaches
It is beyond the scope of this review to consider the limitations of the analysis of scRNA-seq data. As discussed in a recent comprehensive overview of technologies and analytical approaches, there are thousands of informatic tools.74 The approaches used for dimensionality reduction and clustering are crucial. Inappropriate choice of clustering resolutions/parameters can give rise to overclustering or underclustering.74 The potential for overclustering, which may apply to many studies of tissue macrophage heterogeneity, can be addressed by more rigorous statistical analysis of whether clusters constitute truly distinct cell populations.75 Remnant contamination should not be conflated with genuine doublets, which are also a major issue in the analysis of scRNA-seq data. There are several computational approaches to identify and remove so-called heterotypic doublets made up of unrelated cells. Xi et al76 highlight their limitations. Macrophage remnant contaminants may form a unique subset of what is referred to as ambient mRNA, released from lysed cells and contaminating the transcriptome profile.76 As a generalization, we suggest that remnant contribution should be considered in any data set in which macrophages appear to express lineage-specific transcripts from unrelated cells or in which nonmacrophages appear to express Csf1r.
Emerging alternatives to the predominant clustering-based approaches77,78 may mitigate some of the risk of overclustering of scRNA-seq data. Regardless of the analytical method, there is an absolute need for validation of putative subpopulation markers. In the case of mononuclear phagocytes, cellular indexing of transcriptomes and epitopes, in which oligo-conjugated antibodies are used to link cell-surface markers to single-cell transcriptomes (eg, references 27 and 41), or total RNA-seq of populations sorted by flow cytometry3,57 will still be confounded by the remnant issue. Network analysis of very large total RNA-seq data sets from multiple tissues identified clusters of co-regulated transcripts that can provide a reference.3 In principle, any 2 genes that define a subset of macrophages in multiple tissues should be correlated with each other.3 This was clearly not the case for the proposed subsets defined by Lyve1/MHCII/Mrc157 or Lyve1/Folr2/Timd4,12 in which each of the proposed markers clustered independently.3 Many of the available myeloid-specific transgenic reporter lines show evidence of heterogeneous expression in tissue mononuclear phagocytes, but, to date, they have not been used in combination to drive distinct fluorescent reporters.
van Hove et al79 addressed the issue of activation during isolation in a study of brain-associated macrophages. Implementation of a protocol involving the inclusion of actinomycin D to prevent new transcription and reduce digestion temperature (Act-Seq80) reduced but did not eliminate the dissociation artifact. Both cell activation and underrecovery of macrophages (and other cells) in the liver, lung, and other tissues might potentially be addressed using single nucleus RNA-sequencing (snRNA-seq)41,81 or the isolation of active ribosomes using a RiboTag. snRNA-seq does not require the isolation of cells as an intermediate step, so activation is avoided. In a proof-of-concept of the latter approach, a Cx3cr1-cre transgene was used to activate lineage-restricted expression of an epitope-tagged ribosomal subunit gene. Immunoprecipitation of tagged ribosomes from the brain enabled the sequencing of microglia-specific ribosome-associated mRNA.16 In the liver, snRNA-seq retrieved fewer genes per cell but appeared to more accurately reflect the relative abundance of each cell type in vivo.41 Curiously, snRNA-seq applied to mouse lung produced an improved detection of mesenchymal cells but did not appear to increase the relative representation of cells annotated as IM.81
Validation of the existence of subsets of macrophages in cells isolated from any organ would ideally include imaging flow cytometry combined with a reporter transgene/intracellular marker to identify cells in which cytoplasmic and cell-surface markers are discordant.17 Definitive colocalization of 2 markers in situ by immunofluorescence requires tissue clearing and 3-dimensional reconstruction.39,82 Another alternative is to detect the mRNA of interest directly by in situ hybridization, either in the tissue or in isolated cells. For example, we used this approach (with RNAScope) to demonstrate that Csf1r mRNA is expressed only in macrophages in the intestinal lamina propria.83 An alternative technology, Molecular Cartography, which supports multiplex spatial mRNA analysis, was applied to confirm the location of multiple subpopulations of cells in the mouse liver.41 However, densely packed hematopoietic tissues in which macrophage processes often extend multiple cell diameters, including wrapping around other cells, will continue to present challenges with respect to the precision of in situ molecular resolution.
Overview
Macrophages are extremely heterogeneous. However, some of the variation detected with scRNA-seq or other separation technologies on isolated cells does not reflect in vivo biology. Tissue disaggregation recovers only a fraction of resident tissue macrophages, generates remnants that confound much of the analysis, and activates macrophages in the process of isolation. Some genuine variation in macrophage phenotype is associated with the location within subdomains of a tissue, for example, the red pulp and marginal zones of the spleen and the alveolar and interstitial spaces of the lung. Other genuine variations may be a function of life history. What is missing in much of the single-cell data is evidence that markers that define heterogeneity have a function. That question is often addressed by applying a conditional deletion of genes of interest using Cre recombinase, although that approach also has significant potential for artifacts.7 Taking an extreme view, individual macrophages may be almost infinitely heterogeneous,5 yet when we look at their regular distribution in tissues using a Csf1r reporter transgene, they appear remarkably homogeneous.2 We take the view that heterogeneity, per se, could provide an adaptive feature of innate immunity that ensures that each macrophage presents a unique challenge to potential pathogens.
Acknowledgments
The authors are grateful for core support from The Mater Foundation. D.A.H. holds an L3 Investigator Grant (2009750) from the National Health and Medical Research Council (Australia).
Authorship
Contribution: All authors contributed to writing and editing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Hume, Translational Research Institute, Mater Research Institute-University of Queensland, Woolloongabba, QLD 4102, Australia; e-mail: david.hume@uq.edu.au.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal