Introduction:
Clonal dendritic cell (DC) aggregates in chronic myelomonocytic leukemia (CMML) are seen in approximately 20-30% patients at diagnosis. Previously, these were thought to be comprised of CD123+ plasmacytoid DCs but our data has suggested that these are heterogenous and comprise of myeloid DCs, monocytes, and myeloid-derived suppressor cells. Furthermore, these aggregates uniformly express immune checkpoints such as indoleamine 2,3-dioxygenase-1 (IDO), associate with an IDO-specific systemic impact (increased trytophan catabolism), and expanded regulatory T cell population in CMML, suggesting a role in inducing immune tolerance. DC aggregates are also associated with increased risk of acute myeloid leukemia (AML) transformation and resistance to hypomethylating agents, highlighting that they are important for disease progression in CMML (Mangaonkar A et al. Blood Adv 2020). We performed this study to decipher the architecture of a CMML DC aggregate to understand the complex inter-cellular interactions, and assess their impact on the adaptive immune system.
Methods:
In this study, we used archival fresh-frozen paraffin-embedded (FFPE) bone marrow biopsy specimens obtained from CMML patients, and isolated monocytes (CD14+ magnetic bead sorting) and lymphocytes (CD14-ve fraction) from untreated CMML and age-matched normal control (NC) peripheral blood mononuclear cells (PBMCs). DCs were cultured from monocytes using a commercial protocol. Spatial architecture of the CMML BM microenvironment was interrogated through multiplex imaging of FFPE samples by co-detection by indexing (CODEX) using 30 markers (CCR6, CD107a, CD11c, CD20, CD21, CD31, CD3e, CD4, CD44, CD45, CD45RO CD56, CD68, CD8, E-Cadherin, FoxP3, HLA-DR, CD14, CD16, CD11b, CD33, CD15, CD66b, CD123, CD303, CD34, CD163 and IDO). Single-cell transcriptomic signatures were analyzed using the 10X single cell Fixed RNA profiling platform, and validated using bulk RNA seq (Illumina stranded mRNA prep) from isolated and cultured cells. Impact of DCs on lymphocytes was assessed via autologous mixed DC-T culture experiments.
Results:
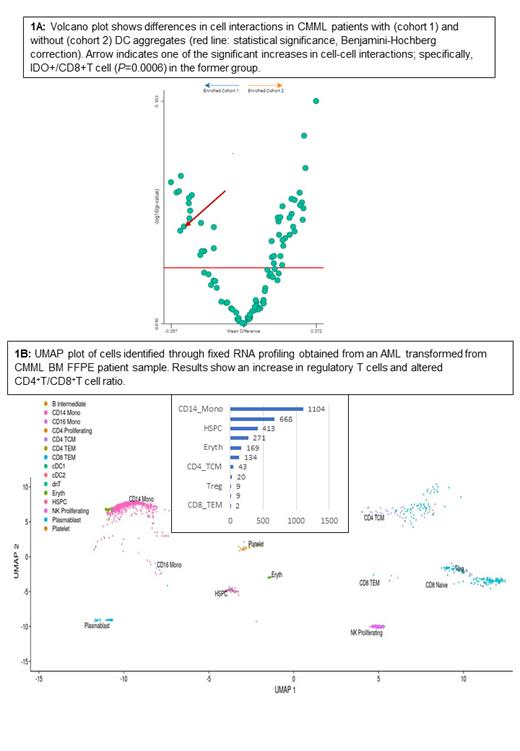
BM FFPE samples from 5 CMML cases (3 with DC aggregates) were included for multiplex imaging via CODEX. Results confirm the significant interactions of CD8+ and CD4+ T cells with CD123+/HLA-DR+ DCs within a CMML DC aggregate (Fig. 1A). Upon quantitative assessment of cell-cell interactions, IDO+/CD8+T ( P=0.006), IDO+/MDSC ( P=0.005), and IDO+/macrophage ( P=0.001) interactions were significantly increased in CMML samples with versus without DC aggregates . Specific findings were validated using single-stain immunohistochemistry and multiplex immunoflourescence staining.
Next, in order to assess transcriptomic signatures within BM at the single-cell level, we chose two cases; CMML (BM aspirate blast% 1, WBC 49.6 x 10 9/L, ASXL1/GATA2/SETBP1/SRSF2 mutations) and AML transformed from CMML (BM aspirate blast% 81, WBC 152 x 10 9/L, NPM1/DNMT3A/TET2 mutations). Approximately, 10,000 cells were obtained and analyzed with cell type annotation through human PBMC reference (Azimuth). Results show an expanded regulatory T cell population in AML compared to CMML (UMAP plot, Fig. 1B), suggestive of immune tolerance.
In order to validate findings and perform functional experiments, we used sorted monocytes and cutured DCs from CMML and NC. DC phenotype was confirmed both through visual inspection and flow cytometry, and clonality was confirmed through Sanger sequencing (for previously identified mutations). Differential gene expression analysis through bulk RNAseq showed significant differences between CMML and NC lymphocytes with immunoinhibitory receptor, LILRB2 (log2fold change=7.05, adjusted P=0.0002) being one of the top five significantly upregulated genes, further confirming immune tolerance. Finally, mixed autologous DC-lymphocyte reaction (1:4 ratio) experiments demonstrated that DCs induce T cell tolerance in CMML (expansion of exhausted PD1/TIM3/LAG3+ CD8 T cells) when compared to NC.
Conclusion: Clonal dendritic cell aggregates within the CMML bone marrow microenviroment interact with T cells and induce immune tolerance. This represents a novel mechanism of disease progression in CMML with potential therapeutic targets (IDO, LILRB2, and/or CD123).
Disclosures
Villasboas:Regeneron: Research Funding; Aptose: Research Funding; Epizyme: Research Funding; Enterome: Research Funding; CRISPR: Research Funding; Genentech: Research Funding. Patnaik:StemLine: Research Funding; CTI Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees; Kura Oncology: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal