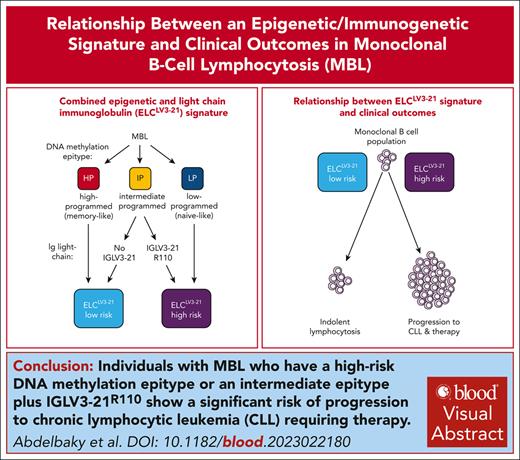
Individuals with MBL who have a high-risk epigenetic/immunogenetic signature show significant risk of progression to CLL requiring therapy.
The signature improves prediction of clinical outcomes compared with other established prognostic indicators regardless of MBL/CLL designation.
Visual Abstract
Monoclonal B-cell lymphocytosis (MBL) progresses to chronic lymphocytic leukemia (CLL) requiring therapy at 1% to 5% per year. Improved prediction of progression would greatly benefit individuals with MBL. Patients with CLL separate into 3 distinct epigenetic subtypes (epitypes) with high prognostic significance, and recently the intermediate epitype has been shown to be enriched for high-risk immunoglobulin lambda variable (IGLV) 3-21 rearrangements, impacting outcomes for these patients. Here, we employed this combined strategy to generate the epigenetic and light chain immunoglobulin (ELCLV3-21) signature to classify 219 individuals with MBL. The ELCLV3-21 high-risk signature distinguished MBL individuals with a high probability of progression (39.9% and 71.1% at 5 and 10 years, respectively). ELCLV3-21 improved the accuracy of predicting time to therapy for individuals with MBL compared with other established prognostic indicators, including the CLL international prognostic index (c-statistic, 0.767 vs 0.668, respectively). Comparing ELCLV3-21 risk groups in MBL vs a cohort of 226 patients with CLL revealed ELCLV3-21 high-risk individuals with MBL had significantly shorter time to therapy (P = .003) and reduced overall survival (P = .03) compared with ELCLV3-21 low-risk individuals with CLL. These results highlight the power of the ELCLV3-21 approach to identify individuals with a higher likelihood of adverse clinical outcome and may provide a more accurate approach to classify individuals with small B-cell clones.
Introduction
Monoclonal B-cell lymphocytosis (MBL) is the emergence of clonal B cells in blood and precedes the manifestation of chronic lymphocytic leukemia (CLL).1-3 MBL is characterized by having a clonal population of B cells at <5 × 109/L in the peripheral blood, with similar immunophenotypic characteristics to CLL and no indication of palpable lymphadenopathy, organomegaly, or cytopenias.4 MBL is divided into low count, if clonal B-cell count is <0.5 × 109/L, and high count, if clonal B-cell count is ≥0.5 × 109/L (subsequently indicated here as MBL). Individuals with MBL are at an increased risk of developing hematologic and non-hematologic cancers,5,6 serious infections,7-9 and progression to CLL requiring therapy, which occurs in 1% to 5% of individuals per year.10,11 As the progression to CLL and the need for therapy are variable, individuals with MBL are likely to experience anxiety and other negative impacts. Improved accuracy to predict risk of clinically adverse outcomes following the identification of MBL would greatly improve the well-being of people identified with MBL and assist in surveillance and management strategies.
Established prognostic markers in CLL, including the mutational status of the immunoglobulin heavy variable (IGHV) locus and the CLL International Prognostic Index (CLL-IPI), have been reported to predict time to first treatment (TTFT) and/or overall survival (OS) among individuals with MBL.12,13 Previously, we and others have published that patients with CLL can be divided into 3 distinct epigenetic subtypes (termed “epitypes”) that reflect progressive DNA methylation changes that occur during B-cell development.14,15 We termed these epitypes low-programmed (LP), intermediate-programmed (IP), and high-programmed (HP) signatures, which predict clinical outcomes irrespective of disease stage and treatment.16,17 Patients with LP-CLL are associated with unfavorable clinical outcomes compared with patients with HP-CLL, whereas patients with IP-CLL display a variable intermediate outcome. Epitype improves prediction of outcomes independent of other well-established prognostic markers, including IGHV mutational status.17
In CLL, unmutated IGHV and IGHV3-21 are associated with inferior outcomes,18,19 and the latter often display IGLV3-21 rearrangements, which also independently portends poor outcomes.20-22 It was subsequently recognized that IGLV3-21 with a p.G110R mutation (termed “R110”), positioned at the variable joining-constant region boundary, generates an autoreactive B-cell receptor configuration and is selected in patients with CLL.21,23 Remarkably, IGLV3-21 use is highly enriched in patients with the IP-CLL epitype, comprising approximately half of total light chain use.17 A landmark study by Nadeu et al22 showed that all IGLV3-21 alleles in patients with the IP-CLL epitype contained the R110 mutation, and that separation of patients with the IP epitype by the presence or absence of the IGLV3-21R110 mutation revealed a similar separation of outcomes as found with poor risk (LP epitype) and favorable risk (HP epitype) groups, respectively. In this study, we applied this approach of combined epitype and IGLV sequencing termed the epigenetic and light chain IGLV3-21 immunoglobulin (ELCLV3-21) signature to classify individuals with MBL.
Study design
Treatment-naïve individuals diagnosed at the Mayo Clinic between 2000 and 2020 with MBL (n = 219) and CLL (n = 226) were identified using the 2018 updated International Workshop on CLL criteria4 (Table 1). To determine ELCLV3-21 status, we first determined the epitype of each individual using the Methylation-iPLEX assay that measures the DNA methylation status of 7 loci to classify individuals into HP, IP, and LP epitypes.17 The λ light chain immunoglobulin rearrangements were determined using Sanger sequencing. We defined the ELCLV3 21 low risk group by combining individuals with the IP epitype (but without IGLV321 rearrangements) and those with the HP epitype. Conversely, we formed the ELCLV321 high risk group by merging individuals with the IP epitype who have IGLV321R110 rearrangements with those in the LP epitype (Figure 1B). Additional details are available in Supplemental Information (available on the Blood website). All individuals provided written informed consent for this research whose protocol was approved by the Mayo Clinic Institutional Review Board
Patient characteristics by cohort
| Characteristic . | Cohort with MBL (n = 219) . | Cohort with CLL (n = 226) . |
|---|---|---|
| Age at diagnosis, y | ||
| Median | 65.89 | 63.06 |
| Range | (44.15-86.38) | (37.39-90.92) |
| Sex | ||
| Female | 88 (40.2) | 63 (27.9) |
| Male | 131 (59.8) | 163 (72.1) |
| Rai stage | ||
| Missing | 0 | 1 |
| 0 | 219 (100.0) | 143 (63.6) |
| I | 0 (0.0) | 53 (23.6) |
| II | 0 (0.0) | 16 (7.1) |
| III | 0 (0.0) | 2 (0.9) |
| IV | 0 (0.0) | 11 (4.9) |
| β-2-Microglobulin, μg/mL | ||
| Missing | 9 | 4 |
| Median | 2.06 | 2.44 |
| Range | (1.00-21.50) | (0.20-14.50) |
| IGHVmutation status | ||
| Missing | 21 | 2 |
| Mutated | 150 (75.8) | 114 (50.9) |
| Unmutated | 48 (24.2) | 110 (49.1) |
| High-risk FISH: (11q, 17p) vs other | ||
| Missing | 10 | 7 |
| FISH: none detected, 13q, trisomy 12 | 201 (96.2) | 190 (86.8) |
| FISH: 11q, 17p | 8 (3.8) | 29 (13.2) |
| CLL-IPI risk | ||
| Missing | 31 | 7 |
| Low risk (0-1) | 121 (64.4) | 83 (37.9) |
| Intermediate risk (2-3) | 48 (25.5) | 83 (37.9) |
| High risk (4-6) | 18 (9.6) | 42 (19.2) |
| Very high risk (7-10) | 1 (0.5) | 11 (5.0) |
| Epitype | ||
| Missing | 0 | 3 |
| HP | 142 (64.8) | 92 (41.3) |
| IP | 42 (19.2) | 45 (20.2) |
| LP | 35 (16.0) | 86 (38.6) |
| ELCLV3-21 | ||
| High risk | 43 (19.6) | 102 (45.1) |
| Low risk | 176 (80.4) | 124 (54.9) |
| Characteristic . | Cohort with MBL (n = 219) . | Cohort with CLL (n = 226) . |
|---|---|---|
| Age at diagnosis, y | ||
| Median | 65.89 | 63.06 |
| Range | (44.15-86.38) | (37.39-90.92) |
| Sex | ||
| Female | 88 (40.2) | 63 (27.9) |
| Male | 131 (59.8) | 163 (72.1) |
| Rai stage | ||
| Missing | 0 | 1 |
| 0 | 219 (100.0) | 143 (63.6) |
| I | 0 (0.0) | 53 (23.6) |
| II | 0 (0.0) | 16 (7.1) |
| III | 0 (0.0) | 2 (0.9) |
| IV | 0 (0.0) | 11 (4.9) |
| β-2-Microglobulin, μg/mL | ||
| Missing | 9 | 4 |
| Median | 2.06 | 2.44 |
| Range | (1.00-21.50) | (0.20-14.50) |
| IGHVmutation status | ||
| Missing | 21 | 2 |
| Mutated | 150 (75.8) | 114 (50.9) |
| Unmutated | 48 (24.2) | 110 (49.1) |
| High-risk FISH: (11q, 17p) vs other | ||
| Missing | 10 | 7 |
| FISH: none detected, 13q, trisomy 12 | 201 (96.2) | 190 (86.8) |
| FISH: 11q, 17p | 8 (3.8) | 29 (13.2) |
| CLL-IPI risk | ||
| Missing | 31 | 7 |
| Low risk (0-1) | 121 (64.4) | 83 (37.9) |
| Intermediate risk (2-3) | 48 (25.5) | 83 (37.9) |
| High risk (4-6) | 18 (9.6) | 42 (19.2) |
| Very high risk (7-10) | 1 (0.5) | 11 (5.0) |
| Epitype | ||
| Missing | 0 | 3 |
| HP | 142 (64.8) | 92 (41.3) |
| IP | 42 (19.2) | 45 (20.2) |
| LP | 35 (16.0) | 86 (38.6) |
| ELCLV3-21 | ||
| High risk | 43 (19.6) | 102 (45.1) |
| Low risk | 176 (80.4) | 124 (54.9) |
Data are given as number (percentage) unless otherwise indicated.
FISH, fluorescence in situ hybridization.
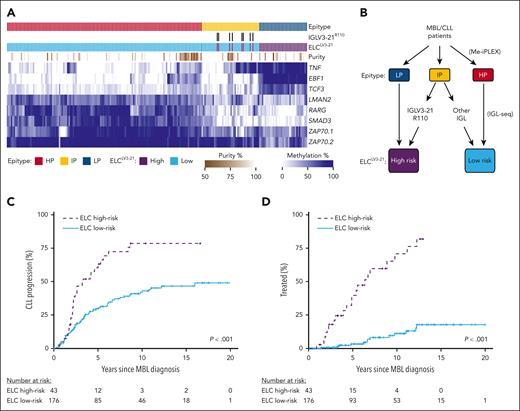
ELCLV3-21 classification of MBL. (A) Heat map displaying DNA methylation of loci used in determining epitype by the CLL Methylation-iPLEX (Me-iPLEX) assay. Epitype random forest classifier and IGLV3-21R110 calls are indicated above. Estimation of sample purity was determined by the Me-iPLEX assay. (B) ELCLV3-21 risk groups were defined by first determining epitype followed by sequencing of immunoglobulin light chain (IGL) gene rearrangements. Individuals with IGLV3-21R110 rearrangements were combined with the LP epitype to generate the ELCLV3-21 high-risk group, and those individuals with light chain rearrangements or lacking the R110 mutation were combined with the HP epitype to produce the ELCLV3-21 low-risk group. (C,D) Cumulative incidence of time to CLL progression (C) and time to first treatment (D) in individuals with MBL separated by ELCLV3-21 risk status.
ELCLV3-21 classification of MBL. (A) Heat map displaying DNA methylation of loci used in determining epitype by the CLL Methylation-iPLEX (Me-iPLEX) assay. Epitype random forest classifier and IGLV3-21R110 calls are indicated above. Estimation of sample purity was determined by the Me-iPLEX assay. (B) ELCLV3-21 risk groups were defined by first determining epitype followed by sequencing of immunoglobulin light chain (IGL) gene rearrangements. Individuals with IGLV3-21R110 rearrangements were combined with the LP epitype to generate the ELCLV3-21 high-risk group, and those individuals with light chain rearrangements or lacking the R110 mutation were combined with the HP epitype to produce the ELCLV3-21 low-risk group. (C,D) Cumulative incidence of time to CLL progression (C) and time to first treatment (D) in individuals with MBL separated by ELCLV3-21 risk status.
Results and discussion
The ELCLV3-21 signature identifies MBL individuals with a high risk of progression to CLL and treatment
We determined 64.8%, 19.2%, and 16.0% of individuals in the MBL cohort to be HP, IP, and LP epitypes, respectively (Figure 1A). We found that 8 of 42 individuals with the IP epitype displayed an IGLV3-21 rearrangement (all showing the R110 mutation) and were used to separate these individuals into ELCLV3-21 risk groups (Figure 1B). Within the MBL group, 105 individuals progressed to CLL, and of these, 34 required therapy. Individuals displaying the IP epitype plus IGLV3-21R110 displayed similar time to CLL progression (TTCP; supplemental Figure 1A) and TTFT (supplemental Figure 1B,C) as other ELCLV3-21 high-risk individuals (LP epitype). To evaluate the longitudinal stability of ELCLV3-21 calls, we remeasured ELCLV3-21 status at later time points (median, 3.7 years; range, 1.2-9.2 years), including after progression to CLL, and observed ELCLV3-21 remained unchanged for 17 of 17 individuals tested (supplemental Table 1). Clonal B-cell counts at diagnosis were not significantly different between ELCLV3-21 groups (P = .31; supplemental Figure 2). ELCLV3-21 high-risk individuals had significantly shorter TTCP (P < .001; Figure 1C) and TTFT (P < .001; Figure 1D), with 39.9% and 71.1% required treatment at 5 and 10 years, respectively. In contrast, among the ELCLV3-21 low-risk individuals, only 3.4% and 11.3% required treatment at 5 and 10 years, respectively.
ELCLV3-21 improves on existing biomarkers for prediction of progression to CLL and treatment among individuals with MBL
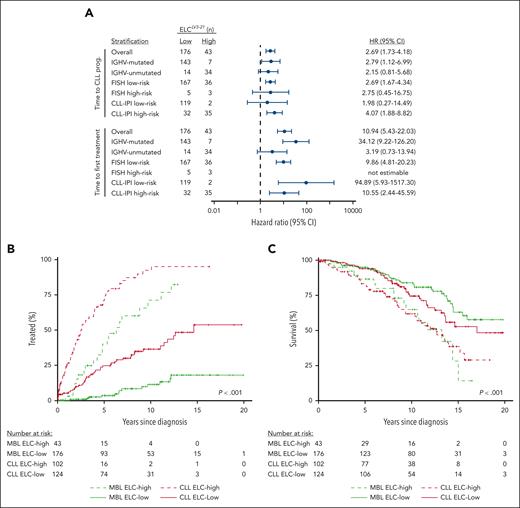
Among common baseline clinical features and CLL prognostic markers, ELCLV3-21 status in individuals with MBL was significantly associated with IGHV mutation status and the CLL-IPI (supplemental Table 2). As IGHV mutation status and CLL-IPI have been previously found to predict TTFT in MBL,13,24 we first stratified individuals with MBL by IGHV status and observed that ELCLV3-21 remained associated with TTCP (hazard ratio [HR], 2.8; 95% confidence interval [CI], 1.1-7.0; P = .03) and TTFT (HR, 34.1; 95% CI, 9.2-126.2; P < .0001) in the IGHV-mutated subgroup (Figure 2A; supplemental Table 3). Among IGHV-unmutated ELCLV3-21, a trend was observed with TTCP (HR, 2.2; 95% CI, 0.8-5.7) and TTFT (HR, 3.2; 95% CI, 0.7-13.9), but did not reach statistical significance (P = .12 each), likely due to smaller numbers in this group. A multivariable model including ELCLV3-21 and IGHV revealed ELCLV3-21 retains significance for TTCP and TTFT, whereas IGHV does not (supplemental Table 4). As the CLL-IPI contains additional features beyond IGHV, we performed multivariable analysis including ELCLV3-21 and CLL-IPI and found that ELCLV3-21 retained significance for TTCP (HR, 3.8; 95% CI, 1.9-7.4; P = .0001) and TTFT (HR, 18.0; 95% CI, 3.3-99.5; P = .009), whereas CLL-IPI did not (supplemental Table 5). ELCLV3-21 improved the accuracy of TTCP and TTFT predictions compared with the CLL-IPI (supplemental Table 6). These findings highlight the importance of ELCLV3-21 beyond well-known markers.
ELCLV3-21 stratifies MBL within other prognostic groups and reveals similar outcomes to CLL patients. (A) Forest plot displaying the hazard ratios of ELCLV3-21 high vs low risk in MBL following stratification for IGHV mutation status, fluorescence in situ hybridization, and CLL-IPI markers in time to first treatment and time to CLL progression. (B,C) Analysis of the impact of ELCLV3-21 in MBL (green lines) vs CLL (red lines) comparing the cumulative incidence of first treatment (B) and overall survival (C) between groups.
ELCLV3-21 stratifies MBL within other prognostic groups and reveals similar outcomes to CLL patients. (A) Forest plot displaying the hazard ratios of ELCLV3-21 high vs low risk in MBL following stratification for IGHV mutation status, fluorescence in situ hybridization, and CLL-IPI markers in time to first treatment and time to CLL progression. (B,C) Analysis of the impact of ELCLV3-21 in MBL (green lines) vs CLL (red lines) comparing the cumulative incidence of first treatment (B) and overall survival (C) between groups.
Individuals with MBL with ELCLV3-21 high risk display similar clinical outcomes compared with those with CLL
In the cohort of patients with CLL (Table 1), 124 (55%) and 102 (45%) were determined to be ELCLV3-21 low and high risk, respectively (supplemental Table 7). ELCLV3-21 was strongly associated with TTFT (P < .001) and OS (P = .006) (Figure 2B,C), consistent with previous findings in CLL.22 When stratifying patients with CLL by IGHV mutation status, more adverse fluorescence in situ hybridization status, or CLL-IPI, the effect of ELCLV3-21 remained significantly associated with TTFT for all, except within the CLL-IPI low-risk stratum (supplemental Table 8). A multivariable model including IGHV and ELCLV3-21 revealed ELCLV3-21 retains significance for TTFT, whereas IGHV does not (supplemental Table 9). Comparing with patients with CLL, ELCLV3-21 high-risk individuals with MBL had a significantly shorter TTFT compared with ELCLV3-21 low-risk patients with CLL (median, 6.3 vs 14.7 years; P = .003; Figure 2B). Similarly, ELCLV3-21 high-risk individuals with MBL displayed inferior OS compared with ELCLV3-21 low-risk patients with CLL (65% vs 74% surviving at 10 years; P = .03; Figure 2C). Notably, the slope of the MBL ELCLV3-21 high-risk curve is similar to that of the CLL high-risk curve, suggesting that these individuals with MBL represent the same biological entity (but detected at an earlier disease stage) and that ELCLV3-21 status may be a useful discriminator of generally benign B-cell clones vs those at risk of progression. Indeed, a multivariable analysis of TTFT, including ELCLV3-21 status and clonal B-cell count (MBL vs CLL), revealed that ELCLV3-21 retained significance for TTFT (P < .0001) and displayed a larger HR than clonal B-cell count (HR, 6.6 [95% CI, 4.6-9.4] vs HR, 3.2 [95% CI, 2.1-4.7], respectively) (supplemental Table 10). Likewise, for predicting OS, ELCLV3-21 retained significance (P < .0001), whereas clonal B-cell count did not (P = .27). Collectively, these findings demonstrate that ELCLV3-21 identifies individuals at the MBL stage who behave comparably to higher-risk patients with CLL.
The identification of a precursor condition such as MBL can cause considerable anxiety and presents an uncertain future. Our findings demonstrate that ELCLV3-21 is a powerful predictor of progression from MBL to CLL and to CLL requiring treatment over a period that surpasses life expectancy for most diagnosed individuals. We also show that ELCLV3-21 measured at the identification of MBL is also predictive for OS, similar to findings in CLL.22 Future studies will be needed to examine the impact of ELCLV3-21 in the era of novel therapies. ELCLV3-21 status captures both DNA methylation epitype and immunogenetic information, 2 important biological features with strong associations to clinical outcomes in CLL.14,15,17,20,21 We demonstrate that ELCLV3-21 provides independent prognostic significance compared with other established CLL prognostic markers, including IGHV mutation status and CLL-IPI. Concordance with patient outcomes on grouping IGLV3-21R110 with IGHV-unmutated individuals to generate the high-risk group was not significantly different from ELCLV3-21 high risk (supplemental Table 11), but as this reclassified only a small minority (6.5%) of patients, a larger cohort would be required detect differences. Improved accuracy of identifying those individuals who have a low risk for progression over an extended period vs those who should be more closely monitored greatly benefits individuals and potentially identifies candidates for emerging strategies for early disease interception. DNA methylation epitype assessment is rapid, cost-effective, and adaptable for clinical use using either pyrosequencing or next-generation sequencing appraoches.16,17 Immunoglobulin gene sequencing is done routinely in a clinical setting, and IGLV3-21R110 can also be assessed by flow cytometry.21 We propose that ELCLV3-21 alone or in combination with other established markers should be included in future validation studies to forecast outcomes of individuals identified with elevated clonal B cells irrespective of cell count at diagnosis.
Acknowledgments
The authors greatly thank the study participants for their contributions and continued participation. This research could not be completed without their valuable time and effort. The authors acknowledge and thank John C. Byrd, University of Cincinnati, for reviewing the final manuscript.
This work was supported by the National Institutes of Health, National Cancer Institute grants R01CA235026 and R01CA258465.
Authorship
Contribution: C.C.O., S.L.S., and E.B. performed concept and study design; W.D., P.J.H., S.B., J.R.C., C.M.V., S.A.P., N.E.K., and T.D.S. acquired patient samples; C.C.O., S.A., B.G., K.Y., Y.-Z.W., S.A.P., A.S.P., N.E.K., T.D.S., S.L.S., and E.B. acquired data; K.G.R., S.A., K.Y., H.Y., and S.L.S. performed data analysis; C.C.O., S.A., S.L.S., and E.B. drafted the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher C. Oakes, Division of Hematology, Departments of Internal Medicine and Biomedical Informatics, The Ohio State University–James Comprehensive Cancer Center, 455 OSU CCC/Wiseman Hall, 410 W 12th Ave, Columbus, OH 43210; email: christopher.oakes@osumc.edu; Susan Slager, Divisions of Hematology and Computational Biology, Mayo Clinic, 200 First Street SW, Rochester, MN 55905; email: Slager.susan@mayo.edu; and Esteban Braggio, Health Futures Center, 6161 E. Mayo Blvd, Room 304G, Phoenix, AZ, 85054; email: Braggio.esteban@mayo.edu.
References
Author notes
S.A. and B.G. contributed equally to this study.
E.B., S.L.S., and C.C.O. contributed equally to this study.
For original data, please contact christopher.oakes@osumc.edu.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal