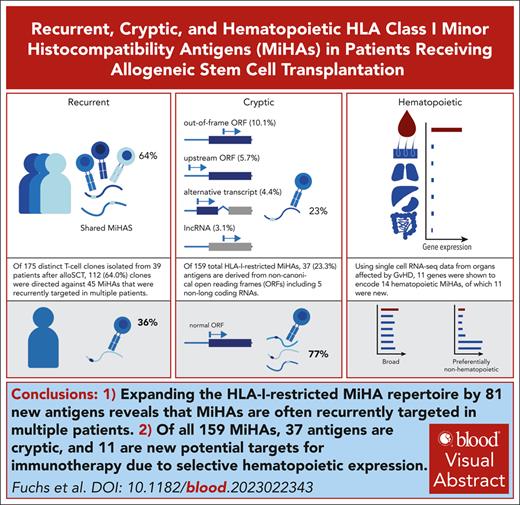
Expanding the HLA-I–restricted MiHA repertoire by 81 new antigens reveals that MiHAs are often recurrently targeted in multiple patients.
Of all 159 MiHAs, 37 antigens are cryptic, and 11 are new potential targets for immunotherapy due to selective hematopoietic expression.
Visual Abstract
Allogeneic stem cell transplantation (alloSCT) is a curative treatment for hematological malignancies. After HLA-matched alloSCT, antitumor immunity is caused by donor T cells recognizing polymorphic peptides, designated minor histocompatibility antigens (MiHAs), that are presented by HLA on malignant patient cells. However, T cells often target MiHAs on healthy nonhematopoietic tissues of patients, thereby inducing side effects known as graft-versus-host disease. Here, we aimed to identify the dominant repertoire of HLA-I-restricted MiHAs to enable strategies to predict, monitor or modulate immune responses after alloSCT. To systematically identify novel MiHAs by genome-wide association screening, T-cell clones were isolated from 39 transplanted patients and tested for reactivity against 191 Epstein-Barr virus transformed B cell lines of the 1000 Genomes Project. By discovering 81 new MiHAs, we more than doubled the antigen repertoire to 159 MiHAs and demonstrated that, despite many genetic differences between patients and donors, often the same MiHAs are targeted in multiple patients. Furthermore, we showed that one quarter of the antigens are cryptic, that is translated from unconventional open reading frames, for example long noncoding RNAs, showing that these antigen types are relevant targets in natural immune responses. Finally, using single cell RNA-seq data, we analyzed tissue expression of MiHA-encoding genes to explore their potential role in clinical outcome, and characterized 11 new hematopoietic-restricted MiHAs as potential targets for immunotherapy. In conclusion, we expanded the repertoire of HLA-I-restricted MiHAs and identified recurrent, cryptic and hematopoietic-restricted antigens, which are fundamental to predict, follow or manipulate immune responses to improve clinical outcome after alloSCT.
Introduction
Allogeneic stem cell transplantation (alloSCT) poses a curative treatment for patients with hematological malignancies. In alloSCT, donor T cells targeting (malignant) hematopoietic cells of patients induce beneficial graft-versus-leukemia (GVL) reactivity, whereas donor T cells targeting patient cells of nonhematopoietic origin cause graft-versus-host disease (GVHD).1 Influencing the balance toward a strong GVL effect while minimizing the risk of GVHD is crucial for therapy outcome. One strategy is to deplete T cells from the graft to reduce incidence and severity of GVHD, followed by delayed donor lymphocyte infusion (DLI) to retain the GVL effect and prevent relapse of disease. However, GVHD remains a serious complication after DLI.2-4
After HLA-matched alloSCT, polymorphic peptides presented on patient cells by HLA surface molecules can be targeted by donor T cells if the peptide is absent in the donor because of genetic differences. The relevance and ambiguous role of these antigens, designated minor histocompatibility antigens (MiHAs), were first recognized when syngeneic transplantations or T cell–depleted transplantations not only led to lower incidence and severity of GVHD, but also increase in relapses.5 Donor T cells targeting broadly expressed MiHAs may not only increase the risk of GVHD, but also contribute to GVL reactivity owing to the presence of MiHAs on malignant cells.6 MiHAs that are exclusively presented on hematopoietic cells, however, constitute safe targets for a selective GVL response without GVHD.7 Since the discovery of the first antigen in 1995,8 78 HLA-I–restricted MiHAs have been identified in patients who underwent transplantation.
Next-generation sequencing identified up to 7500 nonsynonymous single nucleotide polymorphism (SNP) mismatches per patient-donor pair.9,10 These SNPs cause amino acid (AA) changes in annotated open reading frames (ORFs) for proteins, leading to estimated numbers of 50 to 150 polymorphic peptides per HLA allele.11 Although most MiHAs are encoded by nonsynonymous SNPs, other sources of genetic variation also contribute to patient-donor mismatches. For instance, cryptic antigens can be translated from SNPs in coding gene regions in alternative reading frames (out-of-frame ORFs),12-15 SNPs in 5′ untranslated regions (upstream ORFs)15-17 or alternative transcripts.18-22 However, it remains unclear to what extent cryptic antigens contribute to the MiHA repertoire.
Understanding the generation and composition of the MiHA repertoire and its impact on clinical outcome after alloSCT may enable graft manipulation to improve the balance between GVL reactivity and GVHD. Hematopoietic-restricted MiHAs can serve as targets for immunotherapeutic applications, such as adoptive T-cell therapy or vaccine strategies,7,23-25 whereas donor T cells causing GVHD can be depleted. MiHAs may also enable predicting and following clinical outcome or a more directed donor selection with mismatched hematopoietic-restricted antigens and/or matched harmful targets. Therefore, we aimed to expand the current MiHA repertoire using our previously reported genome-wide association screening (GWAS) method optimized for rapid identification of MiHAs in 7 common HLAs (HLA-A∗01:01, A∗02:01, A∗03:01, B∗07:02, B∗08:01, C∗07:01, and C∗07:02).17
In this study, we screened 39 patients who underwent HLA-matched alloSCT and experienced an immune response after DLI. In total, we identified 81 new MiHAs, leading to 159 HLA-I–restricted MiHAs in total. By expanding the repertoire of antigens, we demonstrated that MiHAs are often recurrently targeted in multiple patients. In addition, one quarter of MiHAs were shown to be cryptic antigens, and 11 new MiHAs with hematopoietic-restricted expression were characterized that constitute potential targets for immunotherapeutic interventions.
Methods
Study approval
Peripheral blood and bone marrow mononuclear cells (PBMCs and BMMCs) were collected from patients and donors after approval from the LUMC Institutional Review Board (protocols P03.114, P03.173, and P04.003) and written informed consent according to the Declaration of Helsinki.
Cell culture
T cells were cultured in T-cell medium (TCM: Iscove modified Dulbecco medium [Lonza], 5% human serum [Sanquin], 5% fetal bovine serum [Sigma-Aldrich Chemie], 1.5% glutamine [200 mM], 1% penicillin/streptomycin [200 mM], 0.5 mg/mL amphotericin B, 2 ng/mL interleukin-7 [IL-7] [Miltenyi Biotec], 2 ng/mL IL-15 [Miltenyi Biotec], 120 international unit [IU]/mL IL-2 [Novartis]). Every 2 weeks, T cells were restimulated with 0.8 mg/mL phytohemagglutinin (PHA; Remel Europe) and an irradiated feeder cell mix of third-party PBMCs irradiated with 40 Gy at an E:T ratio of 1:3 to 5 and EBV transformed B-lymphoblastoid cell lines (EBV-LCLs) with 50 Gy at 1:0.3-0.5). EBV-LCLs were cultured in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum, 1.5% glutamine, and 1% penicillin/streptomycin. PHA-activated T cells (PHA-blasts) were generated by adding PHA to PBMCs or BMMCs in TCM. For testing EBV-LCL panels, EBV-LCLs or mixes thereof were cryopreserved in 96-well plates at 60 000 cells per well. Plates were thawed and incubated for 2 days before testing.
T-cell isolation
MiHA-specific T cells were isolated from patient PBMCs after DLI. First, samples were enriched for T cells using a pan T-cell isolation kit (Miltenyi Biotec), and for in vitro stimulation, coincubated for 2 days with irradiated patient PBMCs (15 Gy) obtained before alloSCT in TCM without IL-7 and IL-15, and only 20 IU/ml IL-2. Activated T cells were isolated using fluorescence-activated cell sorter based on fluorescein isothiocyanate–conjugated CD8 (clone RPA-T8, BD/Pharmingen) and allophycocyanin (APC)-conjugated CD137 (clone MOPC-21, BD/Pharmingen) as in vitro marker, or without prior stimulation, based on APC-conjugated HLA-DR (clone G46-6, BD/Pharmingen) as in vivo marker. CD137+ CD8+ cells were also isolated after peptide-stimulation with all MiHAs. Sorted T cells were seeded in 384-well plates at 1, 3, or 10 cells per well and clonally expanded by stimulation with 25 000 and 50 000 irradiated allogeneic PBMCs (40 Gy) per well on day 0 and 7, respectively.
T-cell reactivity assays
Recognition tests were performed in 384-well plates by incubating T cells (2000 cells per well) with EBV-LCLs (15 000 cells per well) or PHA-blasts (30 000 cells per well). For recognition of peptides, donor EBV-LCLs were pulsed with peptides at concentrations from 10 μM to 1 pM or peptide mixes at 10 μM. Supernatant was collected the following day and analyzed using interferon gamma (IFN-γ) enzyme-linked immunosorbent assay (R&D Systems). For recognition of fibroblasts, T cells (5000 cells per well) were incubated with EBV-LCLs (as controls) or fibroblasts (10 000 cells per well) after culturing in absence or presence of 150 IU/mL IFN-γ for 2 days. IFN-γ release was evaluated the following day using enzyme-linked immunosorbent assay (Diaclone).
Identification of MiHAs using GWAS
MiHAs were identified using GWAS as previously described.17 Briefly, T-cell clones were tested against 191 EBV-LCLs sequenced in the 1000 Genomes Project. Based on T-cell reactivity, EBV-LCLs were divided into antigen-positive or -negative if they expressed the HLA shared by recognized EBV-LCLs. To identify MiHA-encoding SNPs associated with T-cell recognition patterns, GWAS was performed using PLINK 1.93.26 Strongly associated SNPs were investigated for encoding polymorphic peptides with predicted HLA-binding using NetMHCpan-4.127 or NetMHCpan-4.0.28 Peptide candidates were validated by T-cell recognition. For identification of HLA-A∗68:01–restricted MiHAs, the EBV-LCL panel was retrovirally transduced with HLA-A∗68:01. For identification of an HY-antigen, T-cell clones were tested against COS-7 cells transfected with HLA-B∗08:01 and 12 genes on the Y chromosome.29,30
Genotyping
Whole-exome sequencing was performed on genomic DNA from patients and donors using QIAmp DNA Micro Kit (Qiagen). For whole-exome capture libraries, Agilent Human All Exon V7 baits were used. Samples were sequenced on Illumina NovaSeq 6000 (PE150) with mean coverage of 50× to generate 150 bp paired-end reads. After filtering using Trimmomatic v0.33 with default parameters (LEADING:15 TRAILING:15 SLIDINGWINDOW:4:15 MINLEN:50), reads were mapped against the GRCh38 reference genome using Burrows-Wheeler Aligner 3 (BWA-mem v0.7.17).31 Duplicate reads were removed using Picard Tools (http://broadinstitute.github.io/picard/). Genome Analysis Toolkit 732 (GATK v4.2.4) was used for base quality recalibration and variant calling. The resulting variant call format (VCF) files for each sample were combined and genotyped using GATK CombineGVCFs and GenotypeGVCFs, respectively. For 15 of 18 SNPs with insufficient coverage, genotyping was performed using KASPar assays (LGC Biosearch Technologies).
Datasets and computational analysis
For frequencies of polymorphic AAs in the human proteome and peptidome, nonsynonymous SNPs retrieved from Ensembl BioMart33 and peptide elution data,34 respectively, were analyzed. For tissue distribution analysis of MiHA-encoding genes, single-cell RNA sequencing data in the Human Protein Atlas (HPA) (v22.0)35 and lab-own Illumina HT12.0 microarray data36 were used. Data were visualized in R using circlize37 and ComplexHeatmap.38
Results
MiHA identification
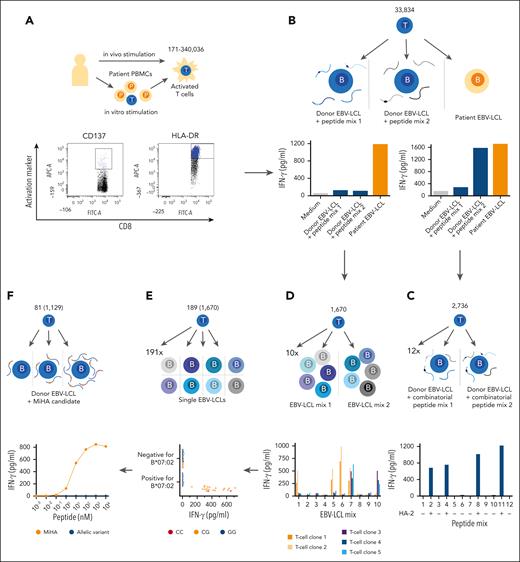
To identify the dominant repertoire of HLA-I–restricted MiHAs, a strategy was followed as outlined in Figure 1. PBMC or BMMC samples were selected at time points after DLI when patients experienced GVHD or conversion from mixed to full donor chimerism. Activated T cells were isolated based on CD137 expression after in vitro stimulation with patient cells obtained before transplantation, or HLA-DR as in vivo activation marker (Figure 1A; supplemental Table 1, available of the Blood website).
Strategy to identify HLA-I-restricted MiHAs. Visual representation of the strategy that was followed to identify HLA-I–restricted MiHAs targeted in 39 patients who responded to DLI after HLA-matched alloSCT with GVL reactivity in the absence or presence of GVHD. (A) Patient samples collected after DLI were enriched for CD8+ T cells. Activated T cells were sorted based on CD137 after 2 days of in vitro stimulation with a pre-alloSCT patient sample (left), or based on HLA-DR as in vivo marker. (B) Growing T-cell clones were tested against patient EBV-LCLs and donor EBV-LCLs pulsed with peptide mixes of known MiHAs. (C) T-cell clones recognizing known antigens were subjected to a combinatorial peptide test to identify the antigen, shown here for the MiHA HA-2. (D) T-cell clones for unknown antigens that exclusively recognized patient EBV-LCL, but not peptide-pulsed donor EBV-LCLs were tested against 10 pools each containing 5 EBV-LCLs. Based on recognition patterns, T-cell clones were clustered, exemplified here by 5 clones with 2 distinct recognition patterns (blue/purple and orange). (E) From each cluster, representative T-cell clones were selected for GWAS. T-cell clones were tested against an optimized panel of 191 EBV-LCLs that were sequenced as part of the 1000 Genomes Project. HLA-restriction was determined by shared HLA alleles on EBV-LCLs that were recognized by T cells (here HLA-B∗07:02-restricted). EBV-LCLs expressing the relevant HLA-restriction allele were divided into antigen-positive and -negative groups based on IFN-γ secretion. All 11.1 million SNPs (minor allele frequency >0.01) were scanned for association with T-cell recognition patterns, here exemplified by heterozygous (orange, nucleotides CG) or homozygous (red, nucleotides CC in example) genotyping for positive EBV-LCLs and homozygous allelic variants (blue, nucleoties GG in example) in negative EBV-LCLs. (F) SNPs with strong association were investigated for encoding polymorphic peptides with predicted binding to the relevant HLA allele, and candidates (orange) and their allelic variants (blue) were tested for T-cell recognition for validation. Numbers in schematic illustration represent range of activated T cells per patients (A), total number of growing T-cell clones (B), T-cell clones for which a MiHA was identified by combinatorial peptide mixes (C), T-cell clones reactive to patient EBV-LCLs (D), T-cell clones for which GWAS performed representative of clustered T-cell clones (number in parentheses) (E) and number of identified new MiHAs (number of T-cell clones targeting these in parentheses) (F).
Strategy to identify HLA-I-restricted MiHAs. Visual representation of the strategy that was followed to identify HLA-I–restricted MiHAs targeted in 39 patients who responded to DLI after HLA-matched alloSCT with GVL reactivity in the absence or presence of GVHD. (A) Patient samples collected after DLI were enriched for CD8+ T cells. Activated T cells were sorted based on CD137 after 2 days of in vitro stimulation with a pre-alloSCT patient sample (left), or based on HLA-DR as in vivo marker. (B) Growing T-cell clones were tested against patient EBV-LCLs and donor EBV-LCLs pulsed with peptide mixes of known MiHAs. (C) T-cell clones recognizing known antigens were subjected to a combinatorial peptide test to identify the antigen, shown here for the MiHA HA-2. (D) T-cell clones for unknown antigens that exclusively recognized patient EBV-LCL, but not peptide-pulsed donor EBV-LCLs were tested against 10 pools each containing 5 EBV-LCLs. Based on recognition patterns, T-cell clones were clustered, exemplified here by 5 clones with 2 distinct recognition patterns (blue/purple and orange). (E) From each cluster, representative T-cell clones were selected for GWAS. T-cell clones were tested against an optimized panel of 191 EBV-LCLs that were sequenced as part of the 1000 Genomes Project. HLA-restriction was determined by shared HLA alleles on EBV-LCLs that were recognized by T cells (here HLA-B∗07:02-restricted). EBV-LCLs expressing the relevant HLA-restriction allele were divided into antigen-positive and -negative groups based on IFN-γ secretion. All 11.1 million SNPs (minor allele frequency >0.01) were scanned for association with T-cell recognition patterns, here exemplified by heterozygous (orange, nucleotides CG) or homozygous (red, nucleotides CC in example) genotyping for positive EBV-LCLs and homozygous allelic variants (blue, nucleoties GG in example) in negative EBV-LCLs. (F) SNPs with strong association were investigated for encoding polymorphic peptides with predicted binding to the relevant HLA allele, and candidates (orange) and their allelic variants (blue) were tested for T-cell recognition for validation. Numbers in schematic illustration represent range of activated T cells per patients (A), total number of growing T-cell clones (B), T-cell clones for which a MiHA was identified by combinatorial peptide mixes (C), T-cell clones reactive to patient EBV-LCLs (D), T-cell clones for which GWAS performed representative of clustered T-cell clones (number in parentheses) (E) and number of identified new MiHAs (number of T-cell clones targeting these in parentheses) (F).
Activated T cells were isolated and growing T-cell clones recognizing patient-derived but not donor-derived target cells were selected (Figure 1B; supplemental Table 1). To identify antigens targeted by T-cell clones recognizing donor EBV-LCLs pulsed with 2 peptide mixes containing previously identified MiHAs, a combinatorial peptide test was performed against 12 peptide mixes with each peptide added to 4 mixes, resulting in a unique recognition pattern (Figure 1C). T-cell clones exclusively recognizing patient cells were assumed to recognize new antigens. Because T-cell clones from the same patient may recognize the same MiHAs, reactivity was tested against 10 mixes of EBV-LCLs (Figure 1D; supplemental Tables 2 and 3). For each patient, T-cell clones with the same recognition pattern were clustered and representative T-cell clones subjected to GWAS.
For identification of SNPs encoding new MiHAs, T-cell clones were tested against 191 EBV-LCLs that were sequenced in the 1000 Genomes Project. GWAS was performed as described previously17 (Figure 1E). SNPs strongly associated with T-cell recognition patterns of EBV-LCLs were investigated for encoding polymorphic HLA-binding peptides. MiHAs were validated by measuring T-cell reactivity against titrated peptides (Figure 1F; supplemental Figure 1; supplemental Table 4). For 1 T-cell clone recognizing an HY-antigen, COS-7 cells expressing HLA-B∗08:01 were transfected with Y chromosome-specific genes,29,30 and predicted HLA-binding peptides encoded by the recognized gene RPS4Y1 were tested for T-cell recognition (supplemental Figure 2).
Patient cohort
T cells were isolated from 53 patients treated with T-cell–depleted, HLA-matched alloSCT followed by prophylactic or pre-emptive DLI. All patients developed immune responses after DLI, defined as conversion to full donor chimerism or GVHD. With 1 exception, samples from patients with GVHD were selected before systemic immunosuppression. Most patients expressed 4 to 6 common HLA-I alleles with population frequencies >20% (HLA-A∗01:01, A∗02:01, A∗03:01, B∗07:02, B∗08:01, C∗07:01, and C∗07:02), for which our GWAS was designed.
MiHAs were successfully identified in 39 patients who developed full donor chimerism without GVHD (n = 11), limited GVHD not requiring systemic immunosuppression (n = 7) or severe GVHD of different grades for affected tissues requiring systemic immunosuppression (n = 21) (Table 1; supplemental Table 5). In 14 patients who converted to full donor chimerism in the absence (n = 8) or presence (n = 6) of GVHD, no MiHAs could be identified potentially because of poor sample quality, activation-induced T-cell death, low MiHA-specific T-cell frequencies, or antigens not presented on EBV-LCLs.
Patient cohort
| . | Total . | No GVHD . | Limited GVHD . | Severe GVHD . |
|---|---|---|---|---|
| Patients | 39 | 11 | 7 | 21 |
| Sex | ||||
| Female | 13 | 4 | 2 | 7 |
| Male | 26 | 7 | 5 | 14 |
| Sex (donor -> patient) | ||||
| f -> f | 12 | 3 | 2 | 7 |
| f -> m | 22 | 4 | 5 | 13 |
| m -> f | 1 | 1 | 0 | 0 |
| m -> m | 4 | 3 | 0 | 1 |
| Relation to donor | ||||
| Unrelated donor | 28 | 6 | 6 | 16 |
| Related donor | 11 | 5 | 1 | 5 |
| Disease | ||||
| AML | 13 | 2 | 3 | 8 |
| B-ALL | 1 | 0 | 1 | 0 |
| CLL | 3 | 1 | 0 | 2 |
| CML | 3 | 2 | 0 | 1 |
| MDS | 7 | 2 | 1 | 4 |
| MM | 8 | 2 | 1 | 5 |
| MPN | 1 | 0 | 0 | 1 |
| t-MN | 1 | 1 | 0 | 0 |
| T-ALL/LBL | 1 | 1 | 0 | 0 |
| T-NHL | 1 | 0 | 1 | 0 |
| Donor/recipient HLA type | ||||
| A∗01:01 | 22 | 7 | 2 | 13 |
| A∗02:01 | 22 | 9 | 5 | 8 |
| A∗03:01 | 16 | 3 | 3 | 10 |
| B∗07:02 | 28 | 9 | 5 | 14 |
| B∗08:01 | 23 | 5 | 3 | 15 |
| C∗07:01 | 25 | 6 | 3 | 16 |
| C∗07:02 | 28 | 9 | 5 | 14 |
| HLA matching | ||||
| 10/10 | 4 | 1 | 2 | 1 |
| 10/12 | 4 | 1 | 0 | 3 |
| 11/12 | 11 | 1 | 1 | 9 |
| 12/12 | 20 | 4 | 8 | 8 |
| . | Total . | No GVHD . | Limited GVHD . | Severe GVHD . |
|---|---|---|---|---|
| Patients | 39 | 11 | 7 | 21 |
| Sex | ||||
| Female | 13 | 4 | 2 | 7 |
| Male | 26 | 7 | 5 | 14 |
| Sex (donor -> patient) | ||||
| f -> f | 12 | 3 | 2 | 7 |
| f -> m | 22 | 4 | 5 | 13 |
| m -> f | 1 | 1 | 0 | 0 |
| m -> m | 4 | 3 | 0 | 1 |
| Relation to donor | ||||
| Unrelated donor | 28 | 6 | 6 | 16 |
| Related donor | 11 | 5 | 1 | 5 |
| Disease | ||||
| AML | 13 | 2 | 3 | 8 |
| B-ALL | 1 | 0 | 1 | 0 |
| CLL | 3 | 1 | 0 | 2 |
| CML | 3 | 2 | 0 | 1 |
| MDS | 7 | 2 | 1 | 4 |
| MM | 8 | 2 | 1 | 5 |
| MPN | 1 | 0 | 0 | 1 |
| t-MN | 1 | 1 | 0 | 0 |
| T-ALL/LBL | 1 | 1 | 0 | 0 |
| T-NHL | 1 | 0 | 1 | 0 |
| Donor/recipient HLA type | ||||
| A∗01:01 | 22 | 7 | 2 | 13 |
| A∗02:01 | 22 | 9 | 5 | 8 |
| A∗03:01 | 16 | 3 | 3 | 10 |
| B∗07:02 | 28 | 9 | 5 | 14 |
| B∗08:01 | 23 | 5 | 3 | 15 |
| C∗07:01 | 25 | 6 | 3 | 16 |
| C∗07:02 | 28 | 9 | 5 | 14 |
| HLA matching | ||||
| 10/10 | 4 | 1 | 2 | 1 |
| 10/12 | 4 | 1 | 0 | 3 |
| 11/12 | 11 | 1 | 1 | 9 |
| 12/12 | 20 | 4 | 8 | 8 |
Patients were selected based on expression of common HLAs and reported GVHD or disappearance of hematopoietic patient cells after DLI.
AML, acute myeloid myeloma; B-ALL, B-cell acute lymphoblastic leukemia, CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; MM, multiple myeloma; MPN, myeloproliferative neoplasm; T-ALL/LBL, T-lymphoblastic leukemia/lymphoma; t-MN, therapy-related myeloid neoplasm; T-NHL, T-cell non-Hodgkin lymphoma.
MiHA repertoire expanded by 81 new antigens
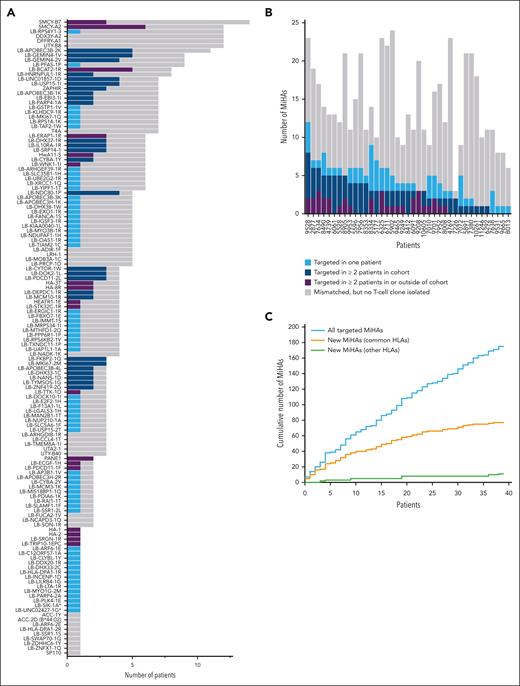
From our cohort of 39 patients, 175 distinct T-cell clones (unique patient-antigen combinations) were isolated for 108 different MiHAs (Figure 2A; supplemental Tables 5 and 6). Of these 108 MiHAs, 32 antigens were previously published. The remaining 76 MiHAs were new. In addition, we applied our optimized GWAS to resolve the target (LB-MKI67-3E) of a T-cell clone from a patient outside our cohort. Because 4 new MiHAs were presented in 2 different HLAs (LB-ARF6-1E/2E and LB-HLA-DPA1-1R/2R in HLA-B∗44:02 & B∗44:03, LB-MYO1G-2M/3M and LB-DHX33-2C/3C in HLA-C∗03:03 & C∗03:04), we identified total 81 new MiHAs (unique peptide-HLA combinations; Table 2), and thus more than doubled the known repertoire to 159 MiHAs (supplemental Table 6).
MiHAs were often recurrently targeted by T cells in 39 patients after HLA-matched alloSCT and DLI. For each MiHA and patient, the number of SNP mismatches were analyzed as well as the number of MiHAs that were targeted as demonstrated by isolation of MiHA-specific T-cell clones. (A) In total, 137 of all 159 MiHAs were mismatched in at least 1 patient that was positive for the relevant HLA-restriction allele (gray). MiHAs were targeted in single patients (light blue) or recurrently targeted in ≥2 patients either in our cohort (dark blue) or also outside our cohort as previously published (purple). Asterisks indicate 2 MiHAs (LB-SIK1-1A and LB-LINC02427-1G) for which T-cell clones were isolated, but SNPs could not be genotyped because of insufficient coverage by whole exome sequencing and KASPar assays. Genotyping also failed for the SNP encoding LB-C16ORF-1R (not displayed). (B) In total, 175 distinct T-cell clones were isolated for a total of 108 different MiHAs. Of the 175 antigen-specific T-cell clones that were isolated, 112 (64.0%) T-cell clones were directed against recurrent antigens that were targeted in ≥2 patients either in our cohort (dark blue) or also outside our cohort (purple). The remaining 63 (36.0%) T-cell clones were directed against MiHAs targeted in only 1 patient (light blue). Indicated are also the total number of MiHA mismatches for each patient with the relevant HLA restriction allele (gray). (C) With each patient added to the study (plotted in order of analysis), the cumulative number of all MiHAs (new and known) that were identified as T-cell targets in each patient increased steadily (blue), while the number of new MiHAs that were identified in common HLAs (orange) stagnated. Similar to all MiHAs, identification of new MiHAs in other HLAs increased (green). New MiHAs were retrospectively assigned to the first patient from whom a specific T-cell clone for the respective antigen was isolated.
MiHAs were often recurrently targeted by T cells in 39 patients after HLA-matched alloSCT and DLI. For each MiHA and patient, the number of SNP mismatches were analyzed as well as the number of MiHAs that were targeted as demonstrated by isolation of MiHA-specific T-cell clones. (A) In total, 137 of all 159 MiHAs were mismatched in at least 1 patient that was positive for the relevant HLA-restriction allele (gray). MiHAs were targeted in single patients (light blue) or recurrently targeted in ≥2 patients either in our cohort (dark blue) or also outside our cohort as previously published (purple). Asterisks indicate 2 MiHAs (LB-SIK1-1A and LB-LINC02427-1G) for which T-cell clones were isolated, but SNPs could not be genotyped because of insufficient coverage by whole exome sequencing and KASPar assays. Genotyping also failed for the SNP encoding LB-C16ORF-1R (not displayed). (B) In total, 175 distinct T-cell clones were isolated for a total of 108 different MiHAs. Of the 175 antigen-specific T-cell clones that were isolated, 112 (64.0%) T-cell clones were directed against recurrent antigens that were targeted in ≥2 patients either in our cohort (dark blue) or also outside our cohort (purple). The remaining 63 (36.0%) T-cell clones were directed against MiHAs targeted in only 1 patient (light blue). Indicated are also the total number of MiHA mismatches for each patient with the relevant HLA restriction allele (gray). (C) With each patient added to the study (plotted in order of analysis), the cumulative number of all MiHAs (new and known) that were identified as T-cell targets in each patient increased steadily (blue), while the number of new MiHAs that were identified in common HLAs (orange) stagnated. Similar to all MiHAs, identification of new MiHAs in other HLAs increased (green). New MiHAs were retrospectively assigned to the first patient from whom a specific T-cell clone for the respective antigen was isolated.
New HLA-I–restricted MiHAs identified by GWAS
| HLA . | MiHA . | Sequence∗ . | SNP . | HLA-allele . | European population allele frequency† . | Type of transcript encoding MiHA . |
|---|---|---|---|---|---|---|
| Common HLAs | ||||||
| A∗01:01 | LB-LINC01857-1D | ST[D/N]ESVLSDY | rs1055228 | A∗01:01 | 0.43 | lncRNA ORF |
| LB-OAS1-1R | ETDDPR[R/T]YQKY | rs1051042 | A∗01:01 | 0.34 | Annotated ORF | |
| LB-SLC35B1-1H | RVD[H/R]TRSWLY | rs1135034 | A∗01:01 | 0.09 | Annotated ORF | |
| LB-UAP1L1-1A | R[A/V]SDGSLLY | rs7037849 | A∗01:01 | 0.60 | Annotated ORF | |
| A∗02:01 | LB-DHX38-1W | ALHY[W/S]DWTC | rs1050361 | A∗02:01 | 0.40 | Out-of-frame ORF |
| LB-E2F2-1H | ALD[H/Q]LIQSC | rs2075995 | A∗02:01 | 0.51 | Annotated ORF | |
| LB-LINC02427-1G | FLWLGAPP[G/S]M | rs1991229 | A∗02:01 | 0.26 | lncRNA ORF | |
| LB-MIS18BP1-1Q | K[Q/E]FPITEAV | rs34402741 | A∗02:01 | 0.01 | Annotated ORF | |
| LB-MTHFD1-1Q | SIIAD[Q/R]IAL | rs2236225 | A∗02:01 | 0.43 | Annotated ORF | |
| LB-NDUFAF1-1H | KLL[H/R]GTYFL | rs1899 | A∗02:01 | 0.27 | Annotated ORF | |
| LB-SLAMF1-1F | GLLSLT[F/L]VL | rs2295612 | A∗02:01 | 0.78 | Annotated ORF | |
| LB-SSR1-2L | VLFRGGPRG[L/S]LAVA | rs10004 | A∗02:01 | 0.75 | Annotated ORF | |
| LB-TIAM2-1C | RL[C/R]KVIQEL | rs11751128 | A∗02:01 | 0.27 | Annotated ORF | |
| A∗03:01 | LB-APOBEC3B-3K | QVYF[K/E]PQYH | rs2076109 | A∗03:01 | 0.40 | Annotated ORF |
| LB-APOBEC3H-2R | [R/G]IFASRLYY | rs139297 | A∗03:01 | 0.46 | Annotated ORF | |
| LB-EXO1-1R | [R/H]SWDDKTCQK | rs735943 | A∗03:01 | 0.57 | Annotated ORF | |
| LB-F13A1-1L | ITFYTGV[L/P]K | rs5982 | A∗03:01 | 0.21 | Annotated ORF | |
| LB-KLHDC9-1R | RLDP[R/S]ARTY | rs11576830 | A∗03:01 | 0.34 | Annotated ORF | |
| LB-MCM10-1R | RA[R/K]GQVLTK | rs2274110 | A∗03:01 | 0.19 | Annotated ORF | |
| LB-NANS-1D | KAL[D/E]RPYTSK | rs1058446 | A∗03:01 | 0.22 | Annotated ORF | |
| LB-SLC5A6-1F | SL[F/L]PLSCQK | rs61737373 | A∗03:01 | 0.07 | Annotated ORF | |
| B∗07:02 | LB-APOBEC3B-4L | TPC[L/P]DCVAKL | rs2076110 | B∗07:02 | 0.06 | Annotated ORF |
| LB-APOBEC3H-1K | KPQQ[K/D]GLRLL | rs139298, rs139299 | B∗07:02 | 0.52 | Annotated ORF | |
| LB-CYTOR-1W | RPLHL[W/R]VVCL | rs7657 | B∗07:02 | 0.45 | lncRNA ORF | |
| LB-DDX20-1R | TPVDD[R/S]ISL | rs197414 | B∗07:02 | 0.87 | Annotated ORF | |
| LB-DHX37-1R | KLASY[R/Q]SCL | rs4447263 | B∗07:02 | 0.40 | Annotated ORF | |
| LB-DOK2-1L | LPRPDSPYSR[L/P] | rs34215892 | B∗07:02 | 0.03 | Annotated ORF | |
| LB-ERGIC1-1R | [R/G]PWPPTLLL | rs477748 | B∗07:02 | 0.29 | Alternative transcript | |
| LB-FANCA-1S | VP[S/G]KYRSLL | rs2239359 | B∗07:02 | 0.41 | Annotated ORF | |
| LB-FBXO7-1E | RPP[E/G]GSGPLL | rs9621461 | B∗07:02 | 0.09 | Out-of-frame ORF | |
| LB-HNRNPUL1-1R | LPSNSR[R/H]HSSL | rs1056854 | B∗07:02 | 0.17 | Out-of-frame ORF | |
| LB-IGSF3-1R | SP[R/Q]DTGNYSC | rs6703791 | B∗07:02 | 0.17 | Annotated ORF | |
| LB-IL10RA-1R | GPRWPP[R/Q]MTH | rs2256111 | B∗07:02 | 0.51 | Out-of-frame ORF | |
| LB-KIAA0040-1L | HPFE[L/P]RTCL | rs1057239 | B∗07:02 | 0.44 | Upstream ORF | |
| LB-LGALS3-1H | KPFKI[H/Q]VL | rs11125 | B∗07:02 | 0.08 | Annotated ORF | |
| LB-LILRB4-1G | [G/D]PRPSPTRSV | rs731170 | B∗07:02 | 0.70 | Annotated ORF | |
| LB-LTA-1R | RV[R/C]GTTLHLLL | rs2229094 | B∗07:02 | 0.29 | Annotated ORF | |
| LB-MKI67-1Q | [Q/R]PRAPRESA | rs10764749 | B∗07:02 | 0.27 | Annotated ORF | |
| LB-MRPS34-1I | RPRDSQ[I/L]YA | rs11552431 | B∗07:02 | 0.13 | Annotated ORF | |
| LB-MYO3B-1R | AP[R/H]SGWPLRSL | rs2161916 | B∗07:02 | 0.44 | Out-of-frame ORF | |
| LB-NUP210-1A | [A/V]PVYTSPQL | rs6795271 | B∗07:02 | 0.68 | Annotated ORF | |
| LB-PDCD11-2L | GPDSSKT[L/F]LCL | rs2986014 | B∗07:02 | 0.58 | Annotated ORF | |
| LB-PFAS-1P | A[P/S]GHTRRKL | rs9891699 | B∗07:02 | 0.17 | Annotated ORF | |
| LB-PPP6R1-1P | MPWV[P/L]MSPF | rs3745920 | B∗07:02 | 0.34 | Out-of-frame ORF | |
| LB-RPS6KB2-1V | SPR[V/A]PVSPLKF | rs13859 | B∗07:02 | 0.42 | Annotated ORF | |
| LB-SIK1-1A | [A/V]PASRASRGGL | rs430554 | B∗07:02 | 0.12 | Annotated ORF | |
| LB-SRP14-1 | LPMGRRRSALW | rs1059395 | B∗07:02 | 0.18 | Out-of-frame ORF | |
| LB-TXNDC11-1P | R[P/L]RGLRLPQL | rs3743588 | B∗07:02 | 0.74 | Out-of-frame ORF | |
| LB-TYMSOS-1G | RPLP[G/R]RIEV | rs2853533 | B∗07:02 | 0.15 | lncRNA ORF | |
| LB-UBE2G2-1R | [R/G]PRVAGGLSVC | rs7364120 | B∗07:02 | 0.29 | Alternative transcript | |
| LB-USP15-1I | MPSHLRN[I/T]LL | rs11174420 | B∗07:02 | 0.31 | Out-of-frame ORF | |
| LB-USP15-2T | MPSHLRN[T/I]LLM | rs11174420 | B∗07:02 | 0.69 | Out-of-frame ORF | |
| LB-ZNF419-2G | IPR[G/D]SWWVEL | rs2074071 | B∗07:02 | 0.69 | Annotated ORF | |
| B∗08:01 | LB-AP3B1-1V | VPN[V/E]KSGAL | rs6453373 | B∗08:01 | 0.07 | Annotated ORF |
| LB-ARHGEF39-1R | SFP[R/H]EKLLLM | rs2297879 | B∗08:01 | 0.33 | Annotated ORF | |
| LB-C12ORF57-1A | L[A/V]FYRKAPL | rs7965269 | B∗08:01 | 0.70 | Upstream ORF | |
| LB-DEPDC1-1R | HLFE[R/Q]ECL | rs78030459 | B∗08:01 | 0.05 | Out-of-frame ORF | |
| LB-MKI67-2M | TP[M/V]QKLDL | rs7918199 | B∗08:01 | 0.08 | Annotated ORF | |
| LB-PARP4-1A | QI[A/T]LHALSL | rs2275660 | B∗08:01 | 0.22 | Annotated ORF | |
| LB-PDIA6-1K | QTKG[K/R]VKL | rs4807 | B∗08:01 | 0.74 | Annotated ORF | |
| LB-RPS14-1K | FLA[K/R]KPSAV | rs4841 | B∗08:01 | 0.26 | Out-of-frame ORF | |
| LB-RPS4Y1-3 | LCKVRKITV | Y chromosone | B∗08:01 | 0.50 | Annotated ORF | |
| LB-TAF2-1W | EPL[W/R]RGASL | rs16893214 | B∗08:01 | 0.20 | Upstream ORF | |
| C∗07:01/C∗07:02 | LB-FKBP2-1Q | MRLSWF[Q/R]VL | rs4672 | C∗07:01 | 0.07 | Annotated ORF |
| LB-XRCC1-1Q | RMRRRLPS[Q/R]RYL | rs25487 | C∗07:01 | 0.37 | Annotated ORF | |
| LB-DOCK10-1I | VRRV[I/N]QEEI | rs78257220 | C∗07:02 | 0.10 | Annotated ORF | |
| Other HLAs | LB-MKI67-3E | ES[E/K]SVQRVTR | rs8473 | A∗68:01 | 0.52 | Annotated ORF |
| LB-PLK4-1E | DSNYPTR[E/D]R | rs17012739 | A∗68:01 | 0.32 | Annotated ORF | |
| LB-SRGN-1R | ESSVQGYPT[R/Q]R | rs2229498 | A∗68:01 | 0.15 | Annotated ORF | |
| LB-PARP4-2A | D[A/G]LGVLPAF | rs1050110 | B∗35:01 | 0.42 | Annotated ORF | |
| LB-MCM3-1K | [K/E]EPFSSVEI | rs2230240 | B∗40:01 | 0.29 | Annotated ORF | |
| LB-ARF6-2E | S[E/Q]GPGGGGDW | rs112017635 | B∗44:02 | 0.14 | Upstream ORF | |
| LB-RAI1-1T | AEQGAQV[T/P]F | rs11649804 | B∗44:02 | 0.33 | Annotated ORF | |
| LB-HLA-DPA1-2R | EEFG[R/Q]AFSF | rs1042178 | B∗44:02 | 0.19 | Annotated ORF | |
| LB-ARF6-1E | S[E/Q]GPGGGGDW | rs112017635 | B∗44:03 | 0.14 | Upstream ORF | |
| LB-HLA-DPA1-1R | EEFG[R/Q]AFSF | rs1042178 | B∗44:03 | 0.19 | Annotated ORF | |
| LB-INCENP-1D | EE[D/E]ARRLRW | rs7129085 | B∗44:03 | 0.63 | Annotated ORF | |
| LB-DHX33-2C | YLYEGGIS[C/R] | rs8069315 | C∗03:03 | 0.12 | Annotated ORF | |
| LB-MYO1G-2M | VS[M/V]NPYQEL | rs61739531 | C∗03:03 | 0.23 | Annotated ORF | |
| LB-DHX33-3C | YLYEGGIS[C/R] | rs8069315 | C∗03:04 | 0.12 | Annotated ORF | |
| LB-MYO1G-3M | VS[M/V]NPYQEL | rs61739531 | C∗03:04 | 0.23 | Annotated ORF | |
| HLA . | MiHA . | Sequence∗ . | SNP . | HLA-allele . | European population allele frequency† . | Type of transcript encoding MiHA . |
|---|---|---|---|---|---|---|
| Common HLAs | ||||||
| A∗01:01 | LB-LINC01857-1D | ST[D/N]ESVLSDY | rs1055228 | A∗01:01 | 0.43 | lncRNA ORF |
| LB-OAS1-1R | ETDDPR[R/T]YQKY | rs1051042 | A∗01:01 | 0.34 | Annotated ORF | |
| LB-SLC35B1-1H | RVD[H/R]TRSWLY | rs1135034 | A∗01:01 | 0.09 | Annotated ORF | |
| LB-UAP1L1-1A | R[A/V]SDGSLLY | rs7037849 | A∗01:01 | 0.60 | Annotated ORF | |
| A∗02:01 | LB-DHX38-1W | ALHY[W/S]DWTC | rs1050361 | A∗02:01 | 0.40 | Out-of-frame ORF |
| LB-E2F2-1H | ALD[H/Q]LIQSC | rs2075995 | A∗02:01 | 0.51 | Annotated ORF | |
| LB-LINC02427-1G | FLWLGAPP[G/S]M | rs1991229 | A∗02:01 | 0.26 | lncRNA ORF | |
| LB-MIS18BP1-1Q | K[Q/E]FPITEAV | rs34402741 | A∗02:01 | 0.01 | Annotated ORF | |
| LB-MTHFD1-1Q | SIIAD[Q/R]IAL | rs2236225 | A∗02:01 | 0.43 | Annotated ORF | |
| LB-NDUFAF1-1H | KLL[H/R]GTYFL | rs1899 | A∗02:01 | 0.27 | Annotated ORF | |
| LB-SLAMF1-1F | GLLSLT[F/L]VL | rs2295612 | A∗02:01 | 0.78 | Annotated ORF | |
| LB-SSR1-2L | VLFRGGPRG[L/S]LAVA | rs10004 | A∗02:01 | 0.75 | Annotated ORF | |
| LB-TIAM2-1C | RL[C/R]KVIQEL | rs11751128 | A∗02:01 | 0.27 | Annotated ORF | |
| A∗03:01 | LB-APOBEC3B-3K | QVYF[K/E]PQYH | rs2076109 | A∗03:01 | 0.40 | Annotated ORF |
| LB-APOBEC3H-2R | [R/G]IFASRLYY | rs139297 | A∗03:01 | 0.46 | Annotated ORF | |
| LB-EXO1-1R | [R/H]SWDDKTCQK | rs735943 | A∗03:01 | 0.57 | Annotated ORF | |
| LB-F13A1-1L | ITFYTGV[L/P]K | rs5982 | A∗03:01 | 0.21 | Annotated ORF | |
| LB-KLHDC9-1R | RLDP[R/S]ARTY | rs11576830 | A∗03:01 | 0.34 | Annotated ORF | |
| LB-MCM10-1R | RA[R/K]GQVLTK | rs2274110 | A∗03:01 | 0.19 | Annotated ORF | |
| LB-NANS-1D | KAL[D/E]RPYTSK | rs1058446 | A∗03:01 | 0.22 | Annotated ORF | |
| LB-SLC5A6-1F | SL[F/L]PLSCQK | rs61737373 | A∗03:01 | 0.07 | Annotated ORF | |
| B∗07:02 | LB-APOBEC3B-4L | TPC[L/P]DCVAKL | rs2076110 | B∗07:02 | 0.06 | Annotated ORF |
| LB-APOBEC3H-1K | KPQQ[K/D]GLRLL | rs139298, rs139299 | B∗07:02 | 0.52 | Annotated ORF | |
| LB-CYTOR-1W | RPLHL[W/R]VVCL | rs7657 | B∗07:02 | 0.45 | lncRNA ORF | |
| LB-DDX20-1R | TPVDD[R/S]ISL | rs197414 | B∗07:02 | 0.87 | Annotated ORF | |
| LB-DHX37-1R | KLASY[R/Q]SCL | rs4447263 | B∗07:02 | 0.40 | Annotated ORF | |
| LB-DOK2-1L | LPRPDSPYSR[L/P] | rs34215892 | B∗07:02 | 0.03 | Annotated ORF | |
| LB-ERGIC1-1R | [R/G]PWPPTLLL | rs477748 | B∗07:02 | 0.29 | Alternative transcript | |
| LB-FANCA-1S | VP[S/G]KYRSLL | rs2239359 | B∗07:02 | 0.41 | Annotated ORF | |
| LB-FBXO7-1E | RPP[E/G]GSGPLL | rs9621461 | B∗07:02 | 0.09 | Out-of-frame ORF | |
| LB-HNRNPUL1-1R | LPSNSR[R/H]HSSL | rs1056854 | B∗07:02 | 0.17 | Out-of-frame ORF | |
| LB-IGSF3-1R | SP[R/Q]DTGNYSC | rs6703791 | B∗07:02 | 0.17 | Annotated ORF | |
| LB-IL10RA-1R | GPRWPP[R/Q]MTH | rs2256111 | B∗07:02 | 0.51 | Out-of-frame ORF | |
| LB-KIAA0040-1L | HPFE[L/P]RTCL | rs1057239 | B∗07:02 | 0.44 | Upstream ORF | |
| LB-LGALS3-1H | KPFKI[H/Q]VL | rs11125 | B∗07:02 | 0.08 | Annotated ORF | |
| LB-LILRB4-1G | [G/D]PRPSPTRSV | rs731170 | B∗07:02 | 0.70 | Annotated ORF | |
| LB-LTA-1R | RV[R/C]GTTLHLLL | rs2229094 | B∗07:02 | 0.29 | Annotated ORF | |
| LB-MKI67-1Q | [Q/R]PRAPRESA | rs10764749 | B∗07:02 | 0.27 | Annotated ORF | |
| LB-MRPS34-1I | RPRDSQ[I/L]YA | rs11552431 | B∗07:02 | 0.13 | Annotated ORF | |
| LB-MYO3B-1R | AP[R/H]SGWPLRSL | rs2161916 | B∗07:02 | 0.44 | Out-of-frame ORF | |
| LB-NUP210-1A | [A/V]PVYTSPQL | rs6795271 | B∗07:02 | 0.68 | Annotated ORF | |
| LB-PDCD11-2L | GPDSSKT[L/F]LCL | rs2986014 | B∗07:02 | 0.58 | Annotated ORF | |
| LB-PFAS-1P | A[P/S]GHTRRKL | rs9891699 | B∗07:02 | 0.17 | Annotated ORF | |
| LB-PPP6R1-1P | MPWV[P/L]MSPF | rs3745920 | B∗07:02 | 0.34 | Out-of-frame ORF | |
| LB-RPS6KB2-1V | SPR[V/A]PVSPLKF | rs13859 | B∗07:02 | 0.42 | Annotated ORF | |
| LB-SIK1-1A | [A/V]PASRASRGGL | rs430554 | B∗07:02 | 0.12 | Annotated ORF | |
| LB-SRP14-1 | LPMGRRRSALW | rs1059395 | B∗07:02 | 0.18 | Out-of-frame ORF | |
| LB-TXNDC11-1P | R[P/L]RGLRLPQL | rs3743588 | B∗07:02 | 0.74 | Out-of-frame ORF | |
| LB-TYMSOS-1G | RPLP[G/R]RIEV | rs2853533 | B∗07:02 | 0.15 | lncRNA ORF | |
| LB-UBE2G2-1R | [R/G]PRVAGGLSVC | rs7364120 | B∗07:02 | 0.29 | Alternative transcript | |
| LB-USP15-1I | MPSHLRN[I/T]LL | rs11174420 | B∗07:02 | 0.31 | Out-of-frame ORF | |
| LB-USP15-2T | MPSHLRN[T/I]LLM | rs11174420 | B∗07:02 | 0.69 | Out-of-frame ORF | |
| LB-ZNF419-2G | IPR[G/D]SWWVEL | rs2074071 | B∗07:02 | 0.69 | Annotated ORF | |
| B∗08:01 | LB-AP3B1-1V | VPN[V/E]KSGAL | rs6453373 | B∗08:01 | 0.07 | Annotated ORF |
| LB-ARHGEF39-1R | SFP[R/H]EKLLLM | rs2297879 | B∗08:01 | 0.33 | Annotated ORF | |
| LB-C12ORF57-1A | L[A/V]FYRKAPL | rs7965269 | B∗08:01 | 0.70 | Upstream ORF | |
| LB-DEPDC1-1R | HLFE[R/Q]ECL | rs78030459 | B∗08:01 | 0.05 | Out-of-frame ORF | |
| LB-MKI67-2M | TP[M/V]QKLDL | rs7918199 | B∗08:01 | 0.08 | Annotated ORF | |
| LB-PARP4-1A | QI[A/T]LHALSL | rs2275660 | B∗08:01 | 0.22 | Annotated ORF | |
| LB-PDIA6-1K | QTKG[K/R]VKL | rs4807 | B∗08:01 | 0.74 | Annotated ORF | |
| LB-RPS14-1K | FLA[K/R]KPSAV | rs4841 | B∗08:01 | 0.26 | Out-of-frame ORF | |
| LB-RPS4Y1-3 | LCKVRKITV | Y chromosone | B∗08:01 | 0.50 | Annotated ORF | |
| LB-TAF2-1W | EPL[W/R]RGASL | rs16893214 | B∗08:01 | 0.20 | Upstream ORF | |
| C∗07:01/C∗07:02 | LB-FKBP2-1Q | MRLSWF[Q/R]VL | rs4672 | C∗07:01 | 0.07 | Annotated ORF |
| LB-XRCC1-1Q | RMRRRLPS[Q/R]RYL | rs25487 | C∗07:01 | 0.37 | Annotated ORF | |
| LB-DOCK10-1I | VRRV[I/N]QEEI | rs78257220 | C∗07:02 | 0.10 | Annotated ORF | |
| Other HLAs | LB-MKI67-3E | ES[E/K]SVQRVTR | rs8473 | A∗68:01 | 0.52 | Annotated ORF |
| LB-PLK4-1E | DSNYPTR[E/D]R | rs17012739 | A∗68:01 | 0.32 | Annotated ORF | |
| LB-SRGN-1R | ESSVQGYPT[R/Q]R | rs2229498 | A∗68:01 | 0.15 | Annotated ORF | |
| LB-PARP4-2A | D[A/G]LGVLPAF | rs1050110 | B∗35:01 | 0.42 | Annotated ORF | |
| LB-MCM3-1K | [K/E]EPFSSVEI | rs2230240 | B∗40:01 | 0.29 | Annotated ORF | |
| LB-ARF6-2E | S[E/Q]GPGGGGDW | rs112017635 | B∗44:02 | 0.14 | Upstream ORF | |
| LB-RAI1-1T | AEQGAQV[T/P]F | rs11649804 | B∗44:02 | 0.33 | Annotated ORF | |
| LB-HLA-DPA1-2R | EEFG[R/Q]AFSF | rs1042178 | B∗44:02 | 0.19 | Annotated ORF | |
| LB-ARF6-1E | S[E/Q]GPGGGGDW | rs112017635 | B∗44:03 | 0.14 | Upstream ORF | |
| LB-HLA-DPA1-1R | EEFG[R/Q]AFSF | rs1042178 | B∗44:03 | 0.19 | Annotated ORF | |
| LB-INCENP-1D | EE[D/E]ARRLRW | rs7129085 | B∗44:03 | 0.63 | Annotated ORF | |
| LB-DHX33-2C | YLYEGGIS[C/R] | rs8069315 | C∗03:03 | 0.12 | Annotated ORF | |
| LB-MYO1G-2M | VS[M/V]NPYQEL | rs61739531 | C∗03:03 | 0.23 | Annotated ORF | |
| LB-DHX33-3C | YLYEGGIS[C/R] | rs8069315 | C∗03:04 | 0.12 | Annotated ORF | |
| LB-MYO1G-3M | VS[M/V]NPYQEL | rs61739531 | C∗03:04 | 0.23 | Annotated ORF | |
Polymorphic AAs are shown between brackets. Bold AA is present in the MiHA, whereas the other AA is present in the allelic variant.
Allele frequency of the MiHA-encoding SNP as reported in the 1000 Genomes Project for the European population.
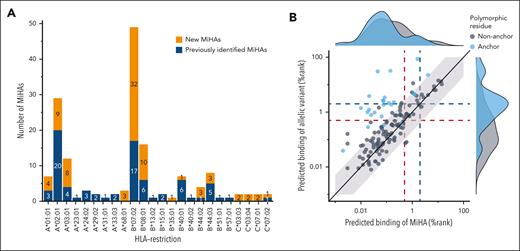
The majority of the 81 new MiHAs were restricted to common HLAs (n = 66), whereas 15 MiHAs were discovered in other HLAs that were expressed by sufficient numbers of EBV-LCLs in the GWAS panel expressing the relevant HLA endogenously (n = 12) or after retroviral transduction (n = 3). The total 159 MiHAs are derived from 129 genes, of which 108 genes encode only 1 MiHA. The other genes produce multiple MiHAs encoded by the same SNP or different SNPs in the same gene, the same peptide in different HLAs or allelic variants in the same HLA (LB-SSR1-1S/2L, LB-PDCD11-1F/2L, LB-USP15-1I/2T, ZAPHIR/LB-ZNF419-2G, HB-1H/Y, and ACC-1C/Y).
To determine how often MiHAs are targeted when mismatched, we genotyped patient-donor pairs for all MiHA-encoding SNPs. In 39 patients, 137 MiHA-encoding SNPs were mismatched in at least 1 patient-donor pair with the relevant HLA. Of these 137 SNP mismatches, 108 MiHAs were shown to be targeted as demonstrated by isolation of specific T-cell clones (Figure 2A; supplemental Figure 3). The most frequently targeted MiHA was SMCY-A2, for which T-cell clones were isolated from 6 of 12 patients mismatched for this MiHA. T-cell clones against LB-FKBP2-1Q and LB-MKI67-2M were each isolated from all 3 patients that were mismatched for these MiHAs, indicating that these antigens are strongly immunogenic. Of the 108 MiHAs targeted in our patient cohort, 45 (41.7%) antigens were recurrently targeted in multiple patients, that is MiHA-specific T-cell clones were isolated from at least 2 patients either both in our cohort or also in patients outside our cohort as previously published. The 45 recurrent MiHAs were targeted by 112 (64.0%) of the 175 isolated T-cell clones (Figure 2B), indicating that immune responses after alloSCT are dominated by recurrent MiHAs.
During our experiments, we kept track of novel MiHAs identified and noticed a gradual saturation in the discovery of MiHAs presented by common HLAs, despite steady increase in the cumulative number of total MiHAs targeted (Figure 2C). This suggests that the dominant repertoire of frequently mismatched MiHAs has been mostly discovered for the common HLAs.
HLA-binding and polymorphic AAs
We next questioned whether MiHAs share biochemical features that lead to higher immunogenicity. Most MiHAs (73.6%; 117 of 159) were presented by 1 of the 7 common HLAs (Figure 3A), in particular HLA-B∗07:02 (30.8%; n = 49) and A∗02:01 (18.2%; n = 29), and exhibited strong HLA-binding predicted by NetMHCpan-4.1 (78.0%; n = 124) (supplemental Table 6; supplemental Figure 4). Other antigens had weak (14.5%; n = 23) or no (7.5%; n = 12) predicted binding. Of 12 MiHAs with no predicted binding, 8 antigens are presented in HLA-A∗02:01. Of all 159 MiHAs, 142 antigens have single polymorphic AAs, in contrast to 17 antigens with multiple polymorphic AAs (LB-TRIP10-1EPC) or peptides that are entirely polymorphic as exemplified by LB-SRP14-1, which is created by a disrupted stop codon. Of the 142 MiHAs with single polymorphic AAs, 25 (17.6%) antigens had polymorphic anchor positions, that is the second (n = 11) or last (n = 11) AA for all HLAs except HLA-B∗08:01 with the third position as anchor residue (n = 3). Most MiHAs with polymorphic anchor residues (80.0%; 20 of 25) had ≥5 times stronger predicted HLA-binding than their allelic counterparts (Figure 3B) compared with only 13 (11.1%) of 117 MiHAs with nonanchor polymorphic AAs. In the remaining 5 MiHAs with polymorphic anchor residues, showing similar predicted HLA-binding as their allelic variants, both AA variants served as anchor residues. The data thus showed that most MiHAs are peptides with predicted HLA-binding in the same range as their allelic variants, suggesting that most allelic variants are presented on the cell surface and that T cells can distinguish MiHAs from their allelic variants by a single AA difference.
MiHAs with polymorphic AAs at nonanchor positions are predicted to bind similarly to HLA as their allelic variants. MiHAs binding to different HLA-I alleles with polymorphic AAs at anchor or nonanchor positions were compared for predicted HLA-binding. (A) The 81 new MiHAs (orange) that were identified by optimized GWAS were mainly found for HLAs for which the method was optimized (HLA-A∗01:01, A∗02:01, A∗03:01, B∗07:02, B∗08:01, C∗07:01, C∗07:02), but also HLAs that were expressed by sufficient numbers of EBV-LCLs in the GWAS panel (HLA-A∗24:02, B∗35:01, B∗40:01, B∗44:02, B∗44:03, C∗03:03) or HLAs that were retrovirally introduced in EBV-LCLs of the GWAS panel (A∗68:01). MiHAs that were previously identified (blue) are also shown. (B) All 142 MiHAs with a single polymorphic AA were divided into 2 groups based on polymorphic residues being at an anchor position (blue) or not (gray). With 20 (80.0%) of 25 MiHAs with a polymorphic anchor residue, the majority of MiHAs showed at least 5 times stronger predicted HLA-binding (outside light gray background) than their allelic counterpart compared to only 13 (11.5%) of 113 MiHAs with a nonanchor polymorphic AA. Indicated are predicted HLA-binding scores by NetMHCpan4.1 for MiHAs (x-axis) and allelic variants (y-axis). High % rank values indicate low predicted HLA-binding. Red and blue dotted lines indicate the cut-offs for strong (% rank <0.5) and weak (% rank <2.0) predicted HLA-binding, respectively.
MiHAs with polymorphic AAs at nonanchor positions are predicted to bind similarly to HLA as their allelic variants. MiHAs binding to different HLA-I alleles with polymorphic AAs at anchor or nonanchor positions were compared for predicted HLA-binding. (A) The 81 new MiHAs (orange) that were identified by optimized GWAS were mainly found for HLAs for which the method was optimized (HLA-A∗01:01, A∗02:01, A∗03:01, B∗07:02, B∗08:01, C∗07:01, C∗07:02), but also HLAs that were expressed by sufficient numbers of EBV-LCLs in the GWAS panel (HLA-A∗24:02, B∗35:01, B∗40:01, B∗44:02, B∗44:03, C∗03:03) or HLAs that were retrovirally introduced in EBV-LCLs of the GWAS panel (A∗68:01). MiHAs that were previously identified (blue) are also shown. (B) All 142 MiHAs with a single polymorphic AA were divided into 2 groups based on polymorphic residues being at an anchor position (blue) or not (gray). With 20 (80.0%) of 25 MiHAs with a polymorphic anchor residue, the majority of MiHAs showed at least 5 times stronger predicted HLA-binding (outside light gray background) than their allelic counterpart compared to only 13 (11.5%) of 113 MiHAs with a nonanchor polymorphic AA. Indicated are predicted HLA-binding scores by NetMHCpan4.1 for MiHAs (x-axis) and allelic variants (y-axis). High % rank values indicate low predicted HLA-binding. Red and blue dotted lines indicate the cut-offs for strong (% rank <0.5) and weak (% rank <2.0) predicted HLA-binding, respectively.
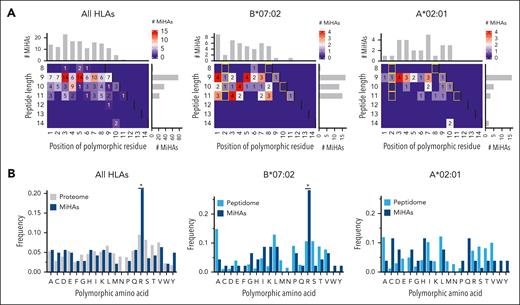
To investigate whether certain residues are more immunogenic than others, we explored their positions and type of polymorphic AAs. Of the 142 MiHAs with single AA changes, polymorphic AAs were observed on almost all positions in 9-, 10- and 11-mer peptides at both anchor and nonanchor residues (Figure 4A). HLA-B∗07:02-binding MiHAs (n = 46) contained similar numbers of 9-, 10- and 11-mer peptides (n = 17, 13, and 15, respectively) with polymorphic AAs predominantly at the first position, whereas HLA-A∗02:01-binding MiHAs (n = 26) were primarily 9-mer peptides (n = 16) with only 1 MiHA with a polymorphic AA on position 1. Arginine was the most frequent polymorphic AA (n = 30; 21.1%). MiHAs with arginine as polymorphic AA were enriched compared with that expected frequencies in the proteome (Figure 4B) and predominantly binding to HLA-B∗07:02 (n = 13), raising the question whether the observed enrichment compared with that in the proteome was due to higher immunogenicity or abundance in the peptidome. We, therefore, analyzed a peptidome data set of polymorphic HLA-B∗07:02- and A∗02:01-binding peptides,34 and showed that HLA-B∗07:02-MiHAs, but not A∗02:01-MiHAs with arginine as polymorphic AA were also enriched compared with the peptidome (Figure 4B). Because predicted binding of HLA-B∗07:02-MiHAs with arginine did not differ from MiHAs with other polymorphic residues (supplemental Figure 5), we concluded that HLA-B∗07:02-peptides with arginine as polymorphic AA are more immunogenic than peptides with other polymorphic AAs.
Position and type of polymorphic AAs in HLA-I-restricted MiHAs. All HLA-I-restricted MiHAs as well as MiHAs binding to HLA-B∗07:02 or HLA-A∗02:01 were analyzed for the position and type of their polymorphic AAs. (A) Polymorphic AAs were observed on anchor (yellow rectangles) and nonanchor positions for all 142 MiHAs with a single polymorphic AA (left) and MiHAs in HLA-B∗07:02 (middle) or HLA-A∗02:01 (right). (B) Of the 142 MiHAs (dark blue), 30 (21.1%) antigens had arginine as polymorphic AA (left panel). This was enriched compared to the expected frequency in the proteome (gray) based on the disparity rate of global allele frequencies of nonsynonymous SNPs in the GRCh38 human genome. For HLA-B∗07:02-binding MiHAs (middle; 13 of 46 (28.3%) antigens), enrichment was also observed compared to the expected frequency of arginine as polymorphic AA in the peptidome (light blue) based on polymorphic peptides eluted from HLA-B∗07:02 (n = 272) on malignant hematopoietic cells and solid tumor cell lines.34 For HLA-A∗02:01-binding MiHAs, no enrichment of arginine as polymorphic AA in comparison to the peptidome (based on 224 polymorphic peptides) was observed. Statistical analysis was performed on the distribution of polymorphic AAs using Spearman rank correlation, and, if statistically significant, post-hoc with 2-sided pairwise binomial test with Bonferroni correction. The difference in distribution of HLA-A∗02:01-peptides was not statistically significant. Asterisks indicate statistically significant (P < .05) differences after Bonferroni correction.
Position and type of polymorphic AAs in HLA-I-restricted MiHAs. All HLA-I-restricted MiHAs as well as MiHAs binding to HLA-B∗07:02 or HLA-A∗02:01 were analyzed for the position and type of their polymorphic AAs. (A) Polymorphic AAs were observed on anchor (yellow rectangles) and nonanchor positions for all 142 MiHAs with a single polymorphic AA (left) and MiHAs in HLA-B∗07:02 (middle) or HLA-A∗02:01 (right). (B) Of the 142 MiHAs (dark blue), 30 (21.1%) antigens had arginine as polymorphic AA (left panel). This was enriched compared to the expected frequency in the proteome (gray) based on the disparity rate of global allele frequencies of nonsynonymous SNPs in the GRCh38 human genome. For HLA-B∗07:02-binding MiHAs (middle; 13 of 46 (28.3%) antigens), enrichment was also observed compared to the expected frequency of arginine as polymorphic AA in the peptidome (light blue) based on polymorphic peptides eluted from HLA-B∗07:02 (n = 272) on malignant hematopoietic cells and solid tumor cell lines.34 For HLA-A∗02:01-binding MiHAs, no enrichment of arginine as polymorphic AA in comparison to the peptidome (based on 224 polymorphic peptides) was observed. Statistical analysis was performed on the distribution of polymorphic AAs using Spearman rank correlation, and, if statistically significant, post-hoc with 2-sided pairwise binomial test with Bonferroni correction. The difference in distribution of HLA-A∗02:01-peptides was not statistically significant. Asterisks indicate statistically significant (P < .05) differences after Bonferroni correction.
Cryptic MiHAs
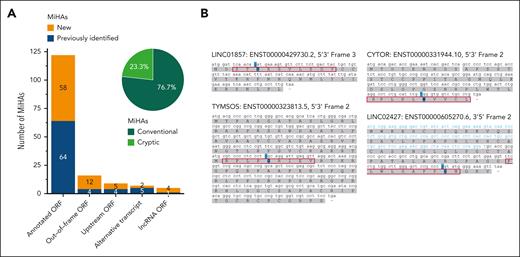
Of all 159 MiHAs, 122 (76.7%) antigens are encoded by missense SNPs in annotated protein-coding ORFs (Figure 5A; supplemental Table 6). The other 37 MiHAs (23.3%) are cryptic peptides from noncanonical ORFs. These cryptic antigens are encoded by missense or synonymous SNPs translated in out-of-frame ORFs (n = 16), upstream ORFs (n = 9), alternative transcripts (n = 7) or long noncoding RNAs (lncRNA ORFs; n = 5). In addition to one previously identified MiHA, which had not been recognized as encoded by an lncRNA ORF,15 we identified 4 new MiHAs encoded by lncRNAs (Figure 5B). Cryptic antigens showed accumulation in HLA-B (31.9%; 29 of 91) compared with that in HLA-A (13.3%; 8 of 60) (supplemental Figure 6). In particular, HLA-B∗07:02 presented many cryptic MiHAs (42.9%; 21 of 49). Our data showed that one quarter of MiHAs are translated in cryptic ORFs, and lncRNA that do not code for proteins with known function, produce relevant targets in natural immune responses after alloSCT.
MiHAs are often translated from noncanonical ORFs. HLA-I–restricted MiHAs were derived from annotated protein-coding reading frames or noncanonical ORFs in normal, alternative, or noncoding transcripts. (A) Of 159 MiHAs (new in orange, previously published in blue), 122 (76.7%) antigens are peptides created by missense SNPs leading to an AA change in canonical ORFs of protein-coding transcripts. This includes 1 antigen in an annotated ORF created by protein splicing. The other 37 (23.3%) antigens are cryptic peptides (light green) created by SNPs causing AA changes in noncanonical ORFs including missense or synonymous SNPs translated in out-of-frame ORFs, SNPs in upstream ORFs, alternative transcripts or lncRNA ORFs. (B) Four new MiHAs are derived from lncRNA ORFs. MiHAs (red boxes) with polymorphic residues (blue) are preceded by start codons. For LINC02427, a start codon is located in the 5′ upstream sequence (blue) of the complementary DNA (black).
MiHAs are often translated from noncanonical ORFs. HLA-I–restricted MiHAs were derived from annotated protein-coding reading frames or noncanonical ORFs in normal, alternative, or noncoding transcripts. (A) Of 159 MiHAs (new in orange, previously published in blue), 122 (76.7%) antigens are peptides created by missense SNPs leading to an AA change in canonical ORFs of protein-coding transcripts. This includes 1 antigen in an annotated ORF created by protein splicing. The other 37 (23.3%) antigens are cryptic peptides (light green) created by SNPs causing AA changes in noncanonical ORFs including missense or synonymous SNPs translated in out-of-frame ORFs, SNPs in upstream ORFs, alternative transcripts or lncRNA ORFs. (B) Four new MiHAs are derived from lncRNA ORFs. MiHAs (red boxes) with polymorphic residues (blue) are preceded by start codons. For LINC02427, a start codon is located in the 5′ upstream sequence (blue) of the complementary DNA (black).
Tissue distribution and potential role in GVHD and GVL
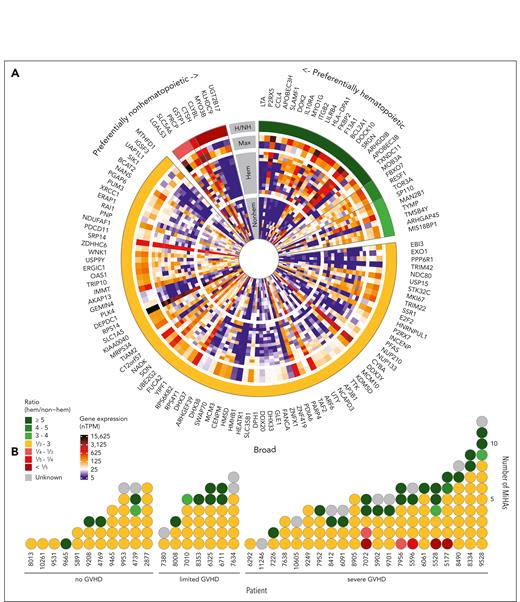
To evaluate the potential impact of MiHAs on clinical outcome, gene expression was analyzed for 123 of 129 MiHA-encoding genes reported in single-cell RNA sequencing data of the HPA (supplemental Table 7).35 Expression in healthy hematopoietic cell clusters was compared with nonhematopoietic cell clusters in organs affected by GVHD, and genes were grouped based on a ratio of maximum expression in hematopoietic compared with nonhematopoietic cell clusters (Figure 6A; supplemental Table 7). Of the 123 genes, 29 (23.5%) genes showed more than or equal to threefold higher expression in hematopoietic cells, whereas 9 (7.3%) genes were more than or equal to threefold higher transcribed in nonhematopoietic cells. The remaining 85 (69.1%) genes were expressed at comparable levels in hematopoietic and nonhematopoietic cells. T-cell clones against MiHAs with preferentially nonhematopoietic expression were only observed in patients with severe GVHD, whereas T-cell clones against MiHAs with preferentially hematopoietic or broad expression were isolated from all patient groups (Figure 6B).
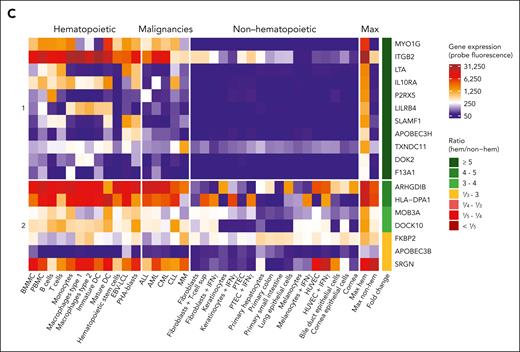
Tissue distribution of MiHAs. Gene expression analysis was performed to distinguish hematopoietic-restricted MiHAs from MiHAs that are broadly expressed or at higher levels in nonhematopoietic tissues. (A) In the single cell section of the HPA, gene expression was reported for 123 of 129 genes coding for 159 MiHAs. Highest gene expression values (nTPM, normalized transcripts per million) for each MiHA in nonhematopoietic (nonhem) and hematopoietic cell clusters (hem) present in tissues involved in GVHD were compared (inner circle: nonhematopoietic cells in skin, esophagus, stomach, small intestine, colon, rectum, liver, lung, bronchus, eye and thymus; second circle: hematopoietic cells including B cells, plasma cells, T cells, natural killer cells, monocytes, dendritic cells, macrophages, Langerhans cells, Kupffer cells, granulocytes, erythroid cells, platelets, and mixed immune cells in GVHD tissues as well as PBMCs and spleen). Cell clusters of undefined mixed cell types were excluded. The maximum expression value in hematopoietic cells (inner lane, third circle) was compared to the maximum value (outer lane, third circle) in nonhematopoietic cell clusters. Genes were grouped based on the calculated ratio (H/NH; outer circle). (B) Indicated is the number of targeted MiHAs in the 39 patients, colored based on their gene expression profiles as defined in panel A. MiHAs with lacking gene expression data are displayed in gray. (C) Microarray data from healthy and malignant hematopoietic cells as well as nonhematopoietic cells cultured under inflammatory conditions were used to analyze gene expression of 20 genes with a ratio ≥5 in hematopoietic cells in the HPA data set. No conclusion could be drawn for 2 genes (CCL4 and BCL2A1) with insufficient probe fluorescence. Of the remaining 18 genes, 11 genes coding for 14 MiHAs showed low expression in nonhematopoietic cell types and therefore have potential as targets for immunotherapy to stimulate GVL reactivity after alloSCT without GVHD (group 1). The other 7 genes showed high expression in ≥1 nonhematopoietic cell types (group 2). DC, dendritic cells; HUVEC, human umbilical vein endothelial cells; PTEC, proximal tubular epithelial cells; sup, supernatant.
Tissue distribution of MiHAs. Gene expression analysis was performed to distinguish hematopoietic-restricted MiHAs from MiHAs that are broadly expressed or at higher levels in nonhematopoietic tissues. (A) In the single cell section of the HPA, gene expression was reported for 123 of 129 genes coding for 159 MiHAs. Highest gene expression values (nTPM, normalized transcripts per million) for each MiHA in nonhematopoietic (nonhem) and hematopoietic cell clusters (hem) present in tissues involved in GVHD were compared (inner circle: nonhematopoietic cells in skin, esophagus, stomach, small intestine, colon, rectum, liver, lung, bronchus, eye and thymus; second circle: hematopoietic cells including B cells, plasma cells, T cells, natural killer cells, monocytes, dendritic cells, macrophages, Langerhans cells, Kupffer cells, granulocytes, erythroid cells, platelets, and mixed immune cells in GVHD tissues as well as PBMCs and spleen). Cell clusters of undefined mixed cell types were excluded. The maximum expression value in hematopoietic cells (inner lane, third circle) was compared to the maximum value (outer lane, third circle) in nonhematopoietic cell clusters. Genes were grouped based on the calculated ratio (H/NH; outer circle). (B) Indicated is the number of targeted MiHAs in the 39 patients, colored based on their gene expression profiles as defined in panel A. MiHAs with lacking gene expression data are displayed in gray. (C) Microarray data from healthy and malignant hematopoietic cells as well as nonhematopoietic cells cultured under inflammatory conditions were used to analyze gene expression of 20 genes with a ratio ≥5 in hematopoietic cells in the HPA data set. No conclusion could be drawn for 2 genes (CCL4 and BCL2A1) with insufficient probe fluorescence. Of the remaining 18 genes, 11 genes coding for 14 MiHAs showed low expression in nonhematopoietic cell types and therefore have potential as targets for immunotherapy to stimulate GVL reactivity after alloSCT without GVHD (group 1). The other 7 genes showed high expression in ≥1 nonhematopoietic cell types (group 2). DC, dendritic cells; HUVEC, human umbilical vein endothelial cells; PTEC, proximal tubular epithelial cells; sup, supernatant.
Because hematopoietic-restricted MiHAs may be relevant targets to stimulate GVL reactivity without GVHD, we further explored 20 genes with more than or equal to fivefold higher expression in hematopoietic cells in our LUMC microarray dataset36 containing hematological malignancies and nonhematopoietic cells treated under inflammatory conditions (Figure 6C; supplemental Table 8). For 2 genes (CCL4 and BCL2A1), expression did not exceed background, suggesting insufficient probe quality. For 7 other genes, high expression was measured in nonhematopoietic cells (ARHGDIB, SRGN, MOB3A, APOBEC3B, DOCK10, and FKBP2) or after culturing under inflammatory conditions (HLA-DPA1). The remaining 11 genes encoding 14 MiHAs were preferentially expressed in hematopoietic cells in both data sets. These MiHAs include 3 antigens previously described as hematopoietic-restricted (HA-2,39,40 LRH-1,18 and LB-ITGB2-121) and 11 new MiHAs that may be relevant for immunotherapy, that is LB-MYO1G-2M, LB-MYO1G-3M, LB-LTA-1R, LB-IL10RA-1R, LB-LILRB4-1G, LB-SLAMF1-1F, LB-APOBEC3H-1K, LB-APOBEC3H-2R, LB-TXNDC11-1P, LB-DOK2-1L, and LB-F13A1-1L.
For 7 of these new MiHAs with hematopoietic-restricted gene expression, reactivity of T-cell clones was tested against skin fibroblasts cultured in the absence or presence of IFN-γ. T-cell clones for LB-APOBEC3H-1K, LB-DOK2-1L, LB-F13A1-1L, LB-IL10RA-1R, LB-LILRB4-1G, LB-LTA-1R, and LB-MYO1G-2M lacked reactivity against fibroblasts (supplemental Figure 7), thereby supporting that the antigens are new hematopoietic-restricted MiHAs.
Discussion
Here, we expanded the repertoire of HLA-I MiHAs by 81 new antigens, thereby more than doubling the repertoire to 159 MiHAs. We demonstrated that the majority of T-cell clones recovered by our methods were directed against a subset of MiHAs that were recurrently targeted in multiple patients and characterized one quarter of MiHAs as cryptic antigens and 11 new hematopoietic-restricted MiHAs with potential therapeutic relevance to stimulate GVL reactivity after alloSCT with a low risk for GVHD.
Up to 12 MiHAs were shown to be targeted in each patient, which may be less than expected considering the high number of SNP mismatches between patient-donor pairs and considerable number of polymorphic HLA-binding peptides on the cell surface.10,11,41 HLA-I–restricted MiHAs were identified by GWAS using EBV-LCLs from the 1000 Genomes Project. By this approach, MiHAs with population frequencies outside detection limits17 and MiHAs expressed on other cell types compared with EBV-LCLs may have been missed. Although T-cell clones for MiHAs with less suitable population frequencies have been isolated, the majority of T-cell clones were directed against MiHAs that were frequently mismatched in patient-donor pairs and successfully identified by GWAS. Furthermore, although it cannot be excluded that MiHAs with expression on other cell types than EBV-LCLs, in particular myeloid cells, may have been missed, efficient immune responses are probably induced by patient-derived professional antigen presenting cells of hematopoietic origin directly presenting MiHAs42,43 and therefore, most activated T cells are expected to react against MiHAs on hematopoietic cells. In a previous study, we specifically searched for T-cell clones that were only reactive against fibroblasts and not patient EBV-LCLs, but were unable to find these T cells in 3 patients with skin GVHD.15 A possible reason could be that T-cell clones were isolated from peripheral blood instead of affected GVHD tissues. T-cell receptor (TCR) sequencing of T cells in blood and affected GVHD tissues showed overlap, but also variation in composition with regards to clonotypes and frequencies. In these studies, however, data interpretation is complicated by limited sampling and information regarding antigen specificities or alloreactivity of the sequenced TCRs.44-46 Koyama et al46 showed that the majority of dominant TCRs found in GVHD-affected skin tissues were also detectable in blood, including the TCR of 1 of 2 confirmed alloreactive T-cell clones. However, Sacirbegovic et al47 showed in mice that TCR repertoires in affected GVHD tissues compared with blood increasingly diverge in time because of T-cell influx from blood in early GVHD and subsequent maintenance by tissue-resident progenitor-like T cells in late GVHD. We often analyzed samples before the onset of clinically diagnosed GVHD, which increases the chance to detect MiHA-specific T cells before homing to tissues.
The low number of MiHAs targeted in each patient may indicate that SNP mismatches often fail to encode polymorphic HLA-binding peptides or encode peptides that are not or weakly immunogenic and dominated in immune responses by other MiHAs, a phenomenon known as clonal dominance.48-50 The observation that often the same MiHAs are targeted in multiple patients suggests similarities among MiHAs in gene expression, protein translation, and peptide processing and presentation, but also presence of high affinity T cells that can recognize and distinguish the antigens from their allelic variants.41,51 Biophysical features of immunogenic peptides have been investigated for shared elements to predict immunogenicity and arginine was described as negatively contributing to immunogenicity in a data set of pathogen, cancer testis, and neoantigen epitopes.52 We observed enrichment of arginine as polymorphic AA, especially in MiHAs binding to HLA-B∗07:02. Although residue preference for arginine has been reported at 3 nonanchor positions for HLA-B∗07:02,53 predicted HLA-B∗07:02-binding was similar between MiHAs with arginine as polymorphic AA and other residues. Moreover, MiHAs with arginine as polymorphic AA were enriched compared with a data set of eluted polymorphic HLA-B∗07:02-peptides. Because MiHAs and their allelic variants often show similar predicted HLA-binding, we speculate that most allelic variants are presented on the cell surface,54-56 and that T cells need to differentiate between both peptides. As such, antigens with arginine as polymorphic AA may be more distinguishable from their allelic variants because of their large positively charged side chain or conformational changes induced in the peptide-HLA-B∗07:02 complex.57
Two predicted indirectly recognizable HLA epitopes (PIRCHEs), that is LB-HLA-DPA1-1R and -2R binding to HLA-B∗44:03 and B∗44:02, respectively, were identified. The T-cell clone for LB-HLA-DPA1-1R was isolated from an HLA-B∗44:03-positive patient after HLA-DP-mismatched alloSCT. In GWAS, the T-cell clone showed recognition of the MiHA on both HLA-B∗44:03- and B∗44:02-positive EBV-LCLs. In previous studies, the number of mismatched HLA-derived epitopes predicted to bind to HLA-I (PIRCHE-I) or HLA-II (PIRCHE-II) were shown to be associated with GVHD and relapse-free survival.58 Based on our findings, we expect a minor contribution of PIRCHE-I as direct targets in CD8 T-cell responses after HLA-matched alloSCT. However, PIRCHE-II antigens may have predictive value for the strength of CD4 T-cell responses after alloSCT. After HLA-matched alloSCT, MiHA-specific CD4 T cells also contribute to clinical responses either by directly targeting HLA-II–positive cells or by stimulating CD8 T-cell responses against HLA-I–restricted MiHAs.59,60
The MiHA repertoire identified by our forward approach (T cell-to-antigen) gives insight in limitations and potentials of reverse (antigen-to-T cell) methods. We found that one quarter of MiHAs are cryptic antigens in noncanonical ORFs. We and others previously identified cryptic antigens translated in out-of-frame ORFs, upstream untranslated regions and alternative transcripts.22,61-65 LncRNAs have also been shown to encode antigens that can be immunogenic upon vaccination in mice66 or in patients with melanoma treated with tumor-infiltrating lymphocytes.67,68 We, here, confirmed that MiHAs translated from lncRNAs are also relevant targets in natural immune responses after alloSCT. Reverse strategies often use proteogenomic approaches to identify HLA-binding peptides by combining whole genome, exome, or transcriptome sequencing with mass spectrometry–based immunopeptidomics. Typically, only nonsynonymous variants leading to AA changes in normal reading frames are included.69-73 To enable identification of cryptic antigens, reference databases need to be enlarged by alternative and long noncoding transcripts and translation of transcripts in all ORFs,65,74-79 which may generate false positives in immunopeptidomics.80 Despite this limitation, reverse strategies estimated that up to 7.5% of HLA-I–associated peptides are noncanonical peptides.77-79 In contrast, we found a higher proportion of 23.3% cryptic MiHAs. Because peptides from cryptic ORFs are similar in peptide length, predicted binding, or hydrophobicity to peptides from canonical ORFs,77-79 no difference in immunogenicity is expected. We, therefore, speculate that for a proportion of cryptic MiHAs, surface expression of the peptide is sufficient for T-cell recognition, but below the detection limit of mass spectrometry.76
Reverse proteogenomic approaches were also used to search for hematopoietic-restricted MiHAs.71,81 Polymorphic HLA-binding peptides were identified by immunopeptidomics and hematopoietic candidates characterized by bulk RNA sequencing analysis. Granados et al81 excluded genes with ubiquitous expression >10 fragments per kilobase million (FPKM) in 27 tissues in the HPA and identified 39 hematopoietic candidates in HLA-A∗02:01 and B∗44:03 with more than or equal to twofold higher expression in bone marrow relative to skin and >1 reads per kilobase million (RPKM) in acute myeloid leukemia in the Cancer Genome Atlas. Olsen et al71 proposed 24 candidates in HLA-A∗02:01, B∗35:01 and C∗07:02 encoded by genes with expression >50 transcripts per million in acute myeloid leukemia in the Cancer Genome Atlas and <50 transcripts per million in nonhematopoietic tissues in the Genotype-Tissue Expression Project. All MiHAs identified by our forward approach were evaluated in a stringent analysis of more than or equal to fivefold higher expression in hematopoietic vs nonhematopoietic cells in 2 independent data sets of single-cell RNA sequencing (HPA) and lab-own microarray data. Our analyses resulted in 14 different hematopoietic-restricted MiHAs which are able to evoke immune responses in patients who underwent transplantation, of which 11 antigens are newly discovered. However, to confirm their therapeutic relevance, T-cell experiments are needed to demonstrate surface presentation of the MiHAs on and killing of malignant hematopoietic cells, while sparing nonhematopoietic cells.
The gradual saturation in MiHA discovery observed during our study indicates that the dominant repertoire of frequently mismatched MiHAs that can be identified by our approach has been mostly characterized for common HLAs. This concise library of MiHAs allows quantification of MiHA-specific T cells in large patient cohorts to understand the relevance of MiHAs for clinical outcome after alloSCT. Previous studies investigated SNP mismatches82 or predicted HLA-binding polymorphic peptides,83-85 but failed to find clear associations with GVHD or GVL. These studies, however, were performed without verification whether the peptides are presented on the cell or able to evoke an immune response. Therefore, the effect of valid MiHAs may have been masked by numerous false-positive peptides. Other studies focused on confirmed MiHAs and showed conflicting data on associations with GVL or GVHD.49,84,86-88 We showed that even for known MiHAs, not every SNP mismatch leads to MiHA-specific T cells in the respective patient as demonstrated by isolation of specific T-cell clones. SMCY-A2, SMCY-B7, and LB-APOBEC3B-2K, for example, were frequently targeted, whereas no T-cell clone was isolated for DDX3-A2, DFFRY-A1, and UTY-B8 though mismatched in a similar number of patients, suggesting hierarchy in immunodominance. MiHAs with broad expression were targeted in patients with presence or absence of GVHD. In patients without GVHD, T-cell frequencies for ubiquitous MiHAs may be lower because of lack of inflammatory cytokines, which are known to stimulate antigen-presentation and T-cell recognition of broad MiHAs on nonhematopoietic cells.15 Therefore, to evaluate immunodominance and estimate the contribution of each MiHA to GVL or GVHD, quantification of MiHA-specific T-cell frequencies in a large patient group is essential.
In conclusion, we expanded the repertoire of HLA-I–restricted MiHAs and identified recurrent, cryptic, and hematopoietic antigens, which are fundamental to predict, follow, or manipulate immune responses to improve clinical outcome after alloSCT.
Acknowledgments
The authors thank all patients and donors for allowing us to use their samples and thereby enabling this research, and the transplantation team of the Department of Hematology (Leiden University Medical Center) for collecting the samples.
This work was supported by the Dutch Cancer Society (project number 10713) and Leiden Center of Computational Oncology. Distribution of EBV-LCLs for the GWAS panel was done under the European Commission seventh Framework Program (FP7) (261123; GEUVADIS).
Authorship
Contribution: K.J.F., J.H.F.F., and M.G. designed the research; K.J.F., M.v.d.M., M.W.H., M.G.D.K., G.K., A.H.d.R., and P.A.v.V. performed research; K.J.F., M.v.d.M., M.W.H., J.H.F.F., and M.G. analyzed and interpreted data; E.A.S.K., C.J.M.H., and P.v.B. curated patient data; J.H.V., C.J.M.H., and P.v.B. organized collection of patient material; I.K. and E.B.v.d.A. contributed to the bioinformatic analysis; K.J.F., C.A.M.v.B., P.A.C.t.H., J.H.F.F., and M.G. wrote the manuscript; J.H.F.F. and M.G. supervised the research; and all authors read and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marieke Griffioen, Department of Hematology, Leiden Medical Research Center, Albinusdreef 2, 2333 ZA Leiden, The Netherlands; email: m.griffioen@lumc.nl.
References
Author notes
Public deposit of antigen sequences to the Immune Epitope Database; for tissue distribution analysis of MiHA-encoding genes, single-cell RNA sequencing data in the Human Protein Atlas (v22.0)35 and lab-own Illumina HT12.0 microarray data36 were used. Data are available on request from the corresponding author, Marieke Griffioen (m.griffioen@lumc.nl).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal