CD22 is expressed in almost all cases of B-cell acute lymphoblastic leukemia (B-ALL) and is therefore an attractive therapeutic target. In this issue of Blood, Zhao and colleagues identified genetic aberrations that impaired clinical response to the CD22-directed antibody-drug conjugate inotuzumab ozogamicin (InO) in adult patients with relapsed or refractory B-ALL.1 Using extensive analysis of pre- and posttreatment patient samples, the authors identified genetic aberrations associated with primary and acquired resistance to InO.
Antibody-drug conjugates are designed for targeted drug delivery: in the case of InO, the antitumor antibiotic calicheamicin is delivered to CD22-positive B cells. Acquired mutations in CD22 were found in 11% of the post-InO relapses, suggesting that CD22-mutated clones were responsible for relapse in some patients. Interestingly, CD22 mutations were not found in baseline samples and did not contribute to resistance in refractory patients. The CD22 mutations led to immune escape by epitope loss (protein truncation, protein destabilization) and epitope alteration. These changes are not always detected by the antibodies used for routine clinical flow cytometry. Beyond the identification of these mutations, Zhao et al modeled the effects of single amino acid changes on the CD22 epitope in the laboratory and confirmed their role in InO escape.
Loss-of-function mutations in TP53 and CDKN2A were present in about one-third of the pre-InO samples and were more common in nonresponders than responders. All 9 patients with a TP53 mutation pre-InO relapsed. Clones with mutations in TP53 and other genes resulting in impaired DNA damage responses emerged after InO treatment, suggesting acquired evasion of InO-induced apoptosis both in relapsed and refractory patients. Interestingly, several patients who relapsed showed a moderate or high number of acquired mutations after InO treatment, and refractory patients or patients who relapsed during InO treatment showed a low number of mutations. Hypermutation was caused by InO-induced DNA damage or tumor intrinsic mismatch repair deficiency and seemed the main driver of CD22 mutation in these cases. Together, DNA damage responses and CD22 mutations explained InO failure in roughly half of the studied cases.
Several earlier reports addressed the topic of resistance to InO (see figure). For example, patients with KMT2A-rearranged and Philadelphia chromosome-positive leukemias had lower response rates, which was associated with lower CD22 expression.2 In pediatric samples, Pennesi et al described calicheamicin resistance as a mechanism of poor response,3 but not CD22 expression, saturation, or internalization of InO after the first dose. Zheng et al described dysregulated CD22 splicing with epitope downregulation as a mechanism of InO resistance.4 Wintering et al reported on the Childhood Oncology Group (COG) experience in study AALL1621 and identified low CD22 with high BCL2 as potential biomarker of poor response.5 Moreover, InO exposure may be correlated with response.6
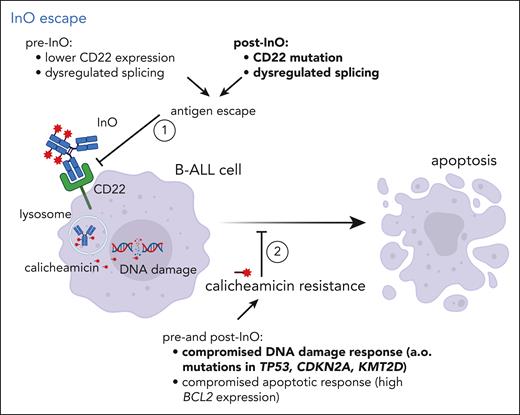
InO escape. Different mechanisms contributing to failure of InO therapy are depicted, particularly (1) antigen escape by alteration of CD22 epitope expression, preventing the antibody-drug conjugate from entering the CD22-positive B cell, and (2) resistance to the drug calicheamicin by compromised DNA damage or apoptotic responses. Mechanisms in bold were observed in the study by Zhao et al.
InO escape. Different mechanisms contributing to failure of InO therapy are depicted, particularly (1) antigen escape by alteration of CD22 epitope expression, preventing the antibody-drug conjugate from entering the CD22-positive B cell, and (2) resistance to the drug calicheamicin by compromised DNA damage or apoptotic responses. Mechanisms in bold were observed in the study by Zhao et al.
InO was approved in adults based on superiority shown in the adult phase 3 InO-VATE trial comparing InO with standard of care: 74% vs 31% complete remission or complete remission with incomplete count recovery (CR/CRi) rate.7 Subsequent studies from MD Anderson developed regimens with alternating blinatumomab and InO on a chemotherapy background of hyper-CVAD (cyclophosphamide, vincristine sulfate, doxorubicin hydrochloride (Adriamycin), dexamethasone, and methotrexate/cytarabine blocks) and including a CD20-directed antibody in CD20-positive cases and showed tolerability and excellent activity of these regimens in adult patients with ALL.8,9 Recently, the Food and Drug Administration approved InO based on single-agent activity in children with CD22-positive relapsed or refractory ALL based on studies of the Innovative Therapies for Children with Cancer (ITCC) consortium.3 Also, in children several studies have now incorporated InO in frontline treatment protocols. With the knowledge gained on InO nonresponse, exposure to InO in earlier phases of treatment may avoid part of the resistance caused by DNA damage response mutations accumulated at (multiple) relapse.
Possible limitations of the study by Zhao et al include its retrospective nature, the limited sample size, and the limited number (28) of paired samples available pre- and post-InO, as well as the heterogeneity in patients and treatment regimens (not all patients received just InO monotherapy). Moreover, the way mutations that do not directly affect the target antigen that may cause resistance was not studied. For example, it could well be that alterations in TP53/DNA damage response genes directly affect the response to chemotherapy, including calicheamicin, or that both resistance mechanisms (epitope escape and chemoresistance) contribute to relapse in the same patient. Moreover, in a large proportion of patients no adequate explanation for poor response was identified, suggesting that other mechanisms are involved.
Based on these findings, it seems premature to implement screening for potential mechanisms of resistance in current clinical practice as flow cytometry would require multiple antibodies to detect CD22 epitope alterations. Another challenge for detection, either by flow or by deep sequencing, is the subclonality of the CD22 mutations resulting in variant allele frequencies per mutation well below 10%. The current study identified CD22 mutations in 11% of post-InO relapse samples; however, it is too early to conclude based on these patient numbers that CD22 mutations could not be present in responders as well. The implications of the identified causes of InO treatment failure may, however, be relevant for patients who would be offered other CD22-targeted therapies, such as chimeric antigen receptor redirected T (CAR-T) cells. Furthermore, some of the findings such as DNA-damage response aberrations and hypermutation may have an impact on escape from other therapies as well.
Most patients treated with InO do respond, which indicates its added value in the growing armamentarium to treat ALL. The correlative biology data presented by Zhao and colleagues show the value of mechanistic studies on biobanked samples. Future prospective studies with InO should incorporate detailed pharmacodynamic studies to better understand the spectrum of resistance.
Conflict-of-interest disclosure: C.M.Z. has received research funding from Syndax, AbbVie, Takeda, Jazz Pharmaceutical, and Pfizer; has been involved as a consultant for BMS, Gilead, Kura Oncology, Novartis, Incyte, and Pfizer; has performed an advisory role for Sanofi and Novartis; and is a member of the board of directors of the ITCC Hem Malignancies Committee. J.M.B. declares no competing financial interest.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal