In this issue of Blood, Simonin et al propose a next-generation sequencing (NGS)-based risk classifier for patients with T-cell acute lymphoblastic leukemia (T-ALL).1 Using an Illumina-based platform sequencing 72 genes (coding exons and adjacent splice junctions), the presence of NOTCH1/FBXW7, PHF6, or EP300 mutations in the absence of N/K-RAS, PI3K pathway (PTEN, PIK3CA, and PIK31R), TP53, DNMT3A, IDH1/2, and IKZF1 mutations confers superior outcomes and classifies patients as low risk (5-year cumulative incidence of relapse [CIR] 21%), and the remaining patients are classified as high risk (5-year CIR 51%). Addition of diagnostic white blood cell (WBC) count and polymerase chain reaction–based end of induction minimal residual disease (MRD) further refines risk stratification, identifying high-, intermediate-, and low-risk groups.
T-ALL is an aggressive leukemia with heterogenous molecular signatures, epigenetic modifications, immunophenotypic diversity, and varying stages of maturation arrest. Unlike B-lineage ALL, for which patients are now risk-stratified based on genomic or cytogenetic alterations, T-ALL has historically eluded molecular risk classification and relied primarily on MRD response to therapy for prognostication, as few features are independently prognostic of disease response. Although outcomes for patients with T-ALL have improved markedly, 15% of children and 30% of adults with T-ALL relapse or have refractory disease, and for these patients, outcomes are dismal.2 Many patients with T-ALL who relapse are classified as low risk at diagnosis. Thus, a critical objective in T-ALL is to improve risk stratification to identify high-risk patients who would most benefit from intensified therapy or precision medicines.
In 2013, the authors of this study proposed a 5-gene risk classifier, demonstrating that mutations in NOTCH1 or FBXW7 in the absence of N/K-RAS mutations or PTEN alterations (mutation or deletion) had significantly more favorable outcomes compared with those with NOTCH1/FBXW7 wild-type or N/K-RAS/PTEN alterations.3 This classifier was replicated in the FRALLE2000T pediatric cohort with similar efficacy.4 However, there have subsequently been mixed results with its application in other cohorts. For the pediatric UKALL2003 trial (which had superior outcomes as compared with FRALLE2000T), the 5-gene classifier had no prognostic impact. However, a post hoc analysis of patients who relapsed demonstrated that the classifier was predictive of outcomes at first relapse.5 Additionally, the 5-gene classifier failed to risk stratify adults treated with a hyper-CVAD (cyclophosphamide, vincristine, adriamycin, dexamethasone) with or without nelarabine regimen, and N/K-RAS and TP53 mutations were highly predictive of poor outcomes.6
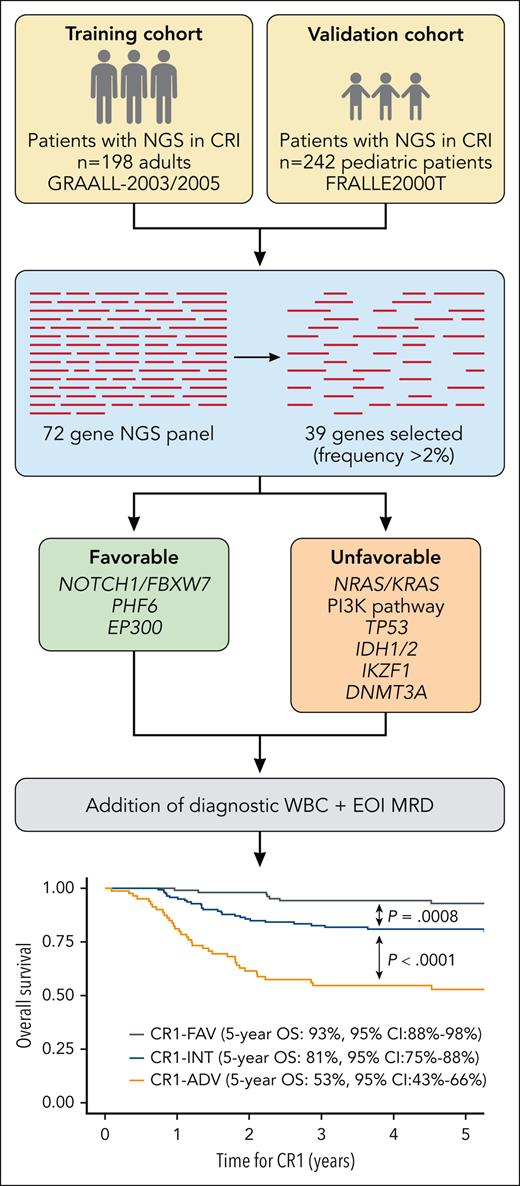
For the current study, the authors revisited the cohorts from which they had derived and replicated the original 5-gene classifier (GRAALL2003/2005 for discovery, FRALLE2000T for validation) and designed a 72-gene NGS panel among patients who achieved complete remission after induction (CR1) with available diagnostic DNA or complementary DNA samples (n = 198 adults, n = 242 children; see figure). The benefits of massively parallel NGS over Sanger sequencing include simultaneous analysis of several hundreds to thousands of genes and greater sensitivity to detect lower-frequency variants.7 NGS is also less costly and is widely used in clinical practice.
Development of an NGS risk classifier in adult and pediatric cohort of patients with T-ALL. CR1-INT, CR1 intermediate risk; EOI, end of induction. Professional illustration by Patrick Lane, ScEYEnce Studios; the graph is reproduced from Figure 4B in the article by Simonin et al that begins on page 1570.
Development of an NGS risk classifier in adult and pediatric cohort of patients with T-ALL. CR1-INT, CR1 intermediate risk; EOI, end of induction. Professional illustration by Patrick Lane, ScEYEnce Studios; the graph is reproduced from Figure 4B in the article by Simonin et al that begins on page 1570.
A LASSO (least absolute shrinkage and selection operator) penalized Fine and Gray model predicting CIR was utilized to select for genes to be retained in the NGS classifier. The primary end point was CIR; secondary end points were disease-free survival and overall survival from CR1 (refractory patients excluded). From the original 72-gene panel, 39 genes were selected to examine for impact on outcomes based on prevalence (affecting >2% of patients). Furthermore, genes within the same pathway were clustered. Ultimately, 4 genes were associated with favorable outcomes (NOTCH1/FBXW7, PHF6, and EP300), and N-K-RAS, PI3K pathway genes, TP53, DNMT3A, IDH1/2, and IKZF1 alterations were significantly associated with higher rates of relapse. Outcomes for individuals in the lowest-risk group, defined by at least 1 favorable mutation and no unfavorable mutations, had a 5-year CIR of 21%. Alternatively, the presence of an adverse gene mutation portended worse outcome, regardless of favorable gene status (5-year CIR 45% with favorable mutation, 53% without favorable mutation). Similarly, unfavorable CIR was observed for patients with absence of favorable or unfavorable alterations, although a notably wide confidence interval suggests heterogeneity within this group (CIR 50%, 95% confidence interval 19%-75%).
To further enhance stratification, the NGS classifier was combined with diagnostic WBC count and end-induction MRD to define 3 risk groups: (1) adverse risk (CR1-ADV) = high-risk NGS-based classifier + high WBC (≥200 × 109 L) or positive MRD (≥10−4); (2) low risk (CR1-FAV)= low-risk NGS-based classifier + low WBC and negative MRD; (3) intermediate risk = all others. This further refined outcomes, particularly in the most extreme groups: 5-year CIR was lower for CR1-FAV patients at 12% and very high for CR1-ADV at 51% (see figure).
Altogether, Simonin et al propose a novel NGS-based risk classification algorithm for T-ALL that successfully stratifies an adult and pediatric cohort. Several of the high-risk genetic alterations have druggable targets, for example, PI3K pathway inhibitors and selective IDH1/IDH2 inhibitors, for which precision medicine approaches may be considered.
Notably, the discovery and validation cohorts for this study are the same 2 cohorts that were initially examined with the Sanger-based 5-gene classifier, and although the original classifier was successful in these cohorts, it was ineffective in other populations. Furthermore, the selection of 39 genes based on frequency of alteration within this European population may minimize applicability across more diverse groups. For example, the frequency of genetic alterations reported herein differs from another sequenced cohort of children or young adults with T-ALL.8 Thus, it remains to be seen if this NGS-based classifier demonstrates portability across modern treatment regimens and among populations of varying ancestral backgrounds. Additionally, the 5-year CIR for “low-risk” patients by the NGS classifier was high (21%), and even with the NGS/WBC/MRD algorithm, relatively high by pediatric standards (12%), leaving room for refinement. Refractory patients were excluded from the analysis. Whether this classifier was effective for this cohort is unreported, and algorithms are needed for these patients, as many patients who relapse are those with refractory disease to induction therapy but who respond to subsequent blocks of therapy, including consolidation/protocol 1B as given on AIEOP-BFM 2000 and COG AALL0434.9,10
Nonetheless, although replication in independent cohorts is critical, this remains an important model for an NGS-based classifier in T-ALL, which could be readily implemented in clinical practice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal