In this issue of Blood, Balducci and colleagues report on the genomic characterization of a large cohort of pediatric (n = 135) and adult (n = 182) patients with T-cell acute lymphoblastic leukemia (T-ALL) derived from the FRALLE2000T and GRAALL03-05 trials using single-nucleotide polymorphism (SNP) microarray and describe novel findings with prognostic relevance.1 Using an assay widely available in the clinical setting, the authors demonstrate that genomic complexity, gain of MLLT4, and deletion of CREBBP were independently predictive of survival in their cohort (see figure). These findings, if validated, may inform risk stratification in future T-ALL clinical trials.
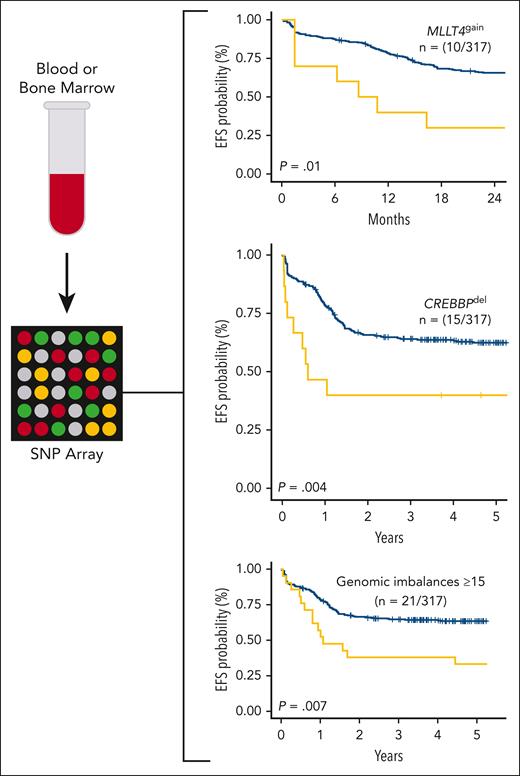
Diagnostic blood or bone marrow samples from patients with T-cell acute lymphoblastic leukemia treated in the FRALLE200T or GRAALL03-05 trials containing ≥70% lymphoblasts were assayed via SNP microarray. Gain of MLLT4, deletion of CREBBP, and ≥15 genomic imbalances were found to be independently prognostic of EFS in a multivariate analysis in the study cohort. EFS, event-free survival; SNP, single-nucleotide polymorphism.
Diagnostic blood or bone marrow samples from patients with T-cell acute lymphoblastic leukemia treated in the FRALLE200T or GRAALL03-05 trials containing ≥70% lymphoblasts were assayed via SNP microarray. Gain of MLLT4, deletion of CREBBP, and ≥15 genomic imbalances were found to be independently prognostic of EFS in a multivariate analysis in the study cohort. EFS, event-free survival; SNP, single-nucleotide polymorphism.
Relapsed T-ALL remains difficult to treat, and prognostic tools to predict relapse are limited. Clinical features, including age, presenting leukocyte count, early T-precursor immunophenotype (ETP-IP), and recurrent genomic alterations, are minimally prognostic in T-ALL. Minimal residual disease (MRD), however, is strongly prognostic and is the only consistent feature currently used for risk stratification across various cooperative groups.2,3 There is an urgent need to identify new prognostic risk factors in T-ALL that are independent of MRD.
The existing literature investigating copy-number alterations in T-ALL is relatively sparse, and the authors enhance this body of knowledge by reporting on the largest cohort to date. Complex karyotype (≥3 cytogenetic alterations) identified by G-banding analysis has been shown to carry an inferior prognosis in adult T-ALL.4,5 However, this technique is limited by reduced sensitivity, a high rate of technical failure (33%),4,5 and the inability to detect cryptic alterations. Prior small studies using SNP arrays in pediatric T-ALL identified a high frequency of CDKN2A loss, a finding replicated by Balducci et al.6,7 However, the authors also identified several novel findings, including a high rate of 13q14 deletions, resulting in loss of RB1/DLEU1 (n = 45; 14%) as well as gain at 6q27 affecting MLLT4 seen in 3% of patients (n = 10). There were relatively minor differences in the pattern of copy-number alterations between the pediatric and adult cohorts, with deletion of CDKN2A occurring more frequently in pediatric cases (79% vs 64%, P = .019) and loss of RB1/DLEU1 and RPL22 found more often in adult cases, although the total number of genomic imbalances per sample did not differ between the cohorts.
The most important aspect of the study is the association of specific copy-number alterations with clinical outcomes. Alterations including chromothripsis, which was only identified in patients with the ETP-IP in this cohort, as well as loss of IKZF1, PTEN, or TP53, were associated with increased relapse risk and/or inferior event-free survival in a univariate analysis. Importantly, genomic complexity (defined as ≥15 genomic imbalances) (n = 21; 6%), gain of MLLT4 (n = 10; 3%), and deletion of CREBBP (n = 15; 5%) were also associated with outcomes in a multivariate analysis that included central nervous system involvement; the presenting leukocyte count; MRD at end-induction; and a previously reported “4-gene” genomic classifier incorporating NOTCH1, FBXW7, RAS, and PTEN alterations.8 Genomic complexity, MLLT4 gain, and CREBBP loss were all independently predictive of a higher relapse rate; MLLT4 gain and CREBBP loss were also associated with inferior event-free survival.
Despite the importance of these findings, there are several limitations that must be considered when determining how to apply the results to T-ALL risk stratification. First, based on the overall sample size, only a few genomic alterations retained prognostic significance in multivariate analyses. Second, several groups were quite rare. For example, only 6 pediatric patients had the ETP-IP. Third, survival analyses considered both the pediatric and adult cohorts together. The significant disparity in outcomes for pediatric and adult T-ALL, as seen in the FRALLE and GRAALL trials, may therefore impact the ability to extrapolate these findings to a purely pediatric or purely adult population. Fourth, the relapse rate in the FRALLE trial (22%) was higher than in other contemporary pediatric T-ALL studies,9 which may similarly limit the generalizability of these results. Fifth, these results need to be confirmed in an independent cohort. For example, the “4-gene” genomic classifier initially described by this group was not validated within a T-ALL cohort from the Medical Research Council,10 limiting its use in other populations. Finally, SNP arrays can only identify certain types of genomic alterations and could miss point mutations or intergenic changes that may impact prognosis.
Despite these concerns, Balducci and colleagues have identified novel genomic markers of prognosis that warrant further investigation in other large T-ALL cohorts. If validated, these results will provide a critical missing element to T-ALL risk stratification---an MRD-independent prognostic variable that can be easily assessed using widely available clinical testing tools that provide results in a clinically meaningful time frame. Such a variable could be used to inform future clinical trial design in T-ALL by identifying patients at high risk for relapse and targeting them for therapy intensification or novel treatment approaches.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal