In this issue of Blood, Morelli et al1 use RNA-targeting CRISPR-Cas13d to reveal that isoforms of long noncoding RNAs (lncRNAs), localized to the nucleus or cytoplasm, are essential for tumor cell survival particularly in multiple myeloma (MM). Notably, their findings are publicly available for the research community through the LongDEP Portal.
lncRNAs are a large family of RNA transcripts exceeding 200 nucleotides in length, characterized by the absence of protein-coding potential. LncRNAs are transcribed from different genomic regions or chromatin states and are known to be localized in distinct subcellular compartments.2 Although these characteristics are critical for defining the potential functions of lncRNAs, the function of the vast majority of identified lncRNAs, and even more so, their different isoforms, remains unknown.3
To investigate the tumor-essential (te) potential of 5327 lncRNA isoforms expressed in patients with newly diagnosed MM, Morelli et al elegantly assessed their essentiality at the RNA level using the CRISPR-Cas13d system in MM cell lines and other human tumors. Unlike Cas9 nucleases, which are commonly used in CRISPR screens for genome editing, or dCas9 (a nuclease-dead Cas9 variant) used to repress transcription via DNA binding, Cas13d, one of the smallest nucleases of the Cas13 family, exclusively targets and cleaves single-stranded RNA without binding or cutting DNA.4 LncRNAs often overlap with or share promoters with coding or noncoding genes and contain regulatory elements such as small RNAs. Notably, lncRNAs generate multiple isoforms, making their precise functional characterization difficult using conventional Cas9 approaches. Cas13d’s ability to target RNA molecules directly is ideal for accurately determining not only which lncRNAs are critical but also which specific isoforms are essential for tumor cell survival.
Using this innovative CRISPR-Cas13d approach, the authors identified 183 te-lncRNA isoforms in MM cell lines and xenografts, 86 of which were found to be essential across other tumor types, thus defining them as common te-lncRNA isoforms in cancer. Thus, this CRISPR-Cas13d guide library is a useful platform for the identification of te-lncRNA isoforms in human tumors. Furthermore, the authors demonstrated that high expression levels of te-lncRNA isoforms correlated with their essentialness in tumor cells, an important observation, since mechanisms such as epigenomic alterations, the activation of alternative promoters,5 or the influence of specific transcription factors6 could drive the elevated expression of particular isoforms in cancer. Future studies exploring these mechanisms will be crucial to better define the functional roles of individual lncRNA isoforms and to understand why some are selectively essential for tumor cell survival.
Beyond uncovering tumor-essential lncRNA isoforms, Morelli et al observed that the high expression of 92 te-lncRNA isoforms correlated with worse overall survival in patients with MM. These results are consistent with previous studies highlighting the prognostic significance of lncRNAs in MM,2,3,7 suggesting that lncRNA expression could aid in risk stratification. Although their incorporation into routine clinical practice remains uncertain in the short term, these results reinforce the growing importance of lncRNAs in the pathobiology of MM.
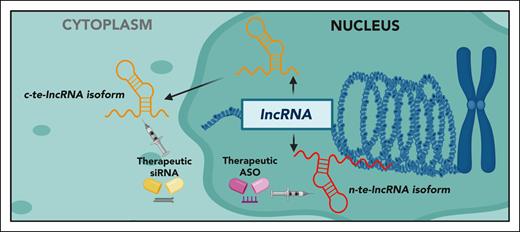
Finally, the authors mapped the subcellular localization of te-lncRNAs isoforms, measuring their expression across chromatin-bound, nuclear-soluble, and cytoplasmic cellular fractions. As expected, the authors observed that the majority of te-lncRNA isoforms exhibited their highest expression in the chromatin-bound fraction. However, the study also revealed that certain te-lncRNA isoforms, such as SNHG6-003 isoform of the lncRNA SNHG6, were predominantly expressed and functionally active in the cytoplasm. Subcellular localization is critical to understanding lncRNA functionality and has major implications for the development of RNA-based therapies. RNA-based treatments, which involve delivering RNA molecules into cells to modulate gene expression, have gained significant attention, particularly after the success of mRNA vaccines for COVID-19, and offer attractive potential for cancer therapy. Determining the subcellular compartment where a specific lncRNA isoform operates enables the design of targeted RNA-based strategies, such as antisense oligonucleotides (ASOs) for nuclear te-lncRNAs or small interfering RNAs (siRNAs) for cytoplasmic te-lncRNAs (see figure). Precision targeting could minimize off-target effects and enhance therapeutic efficacy in cancer cells. Although RNA-based approaches have shown promising results, especially in vitro, further improvements in stability, pharmacokinetics, and tumor-specific delivery are needed. To overcome these challenges, it is critical to develop advanced delivery systems capable of protecting RNA molecules from nuclease degradation while ensuring efficient and selective delivery to target tumor cells.8 Continued progress in this area will be essential for the clinical translation of RNA-based therapies and could ultimately lead to major advances in the treatment of MM and other cancers.
Expression of nuclear and cytoplasmic essential lncRNA isoforms in MM cell. RNA-based therapeutic strategies, such as the use of ASOs targeting nuclear (n) te-lncRNA isoforms or siRNAs targeting cytosolic (c) te-lncRNA isoforms, have the potential to induce antiproliferative effects in MM cells. Figure created with BioRender.com.
Expression of nuclear and cytoplasmic essential lncRNA isoforms in MM cell. RNA-based therapeutic strategies, such as the use of ASOs targeting nuclear (n) te-lncRNA isoforms or siRNAs targeting cytosolic (c) te-lncRNA isoforms, have the potential to induce antiproliferative effects in MM cells. Figure created with BioRender.com.
Altogether, this study not only expands our understanding of the functional landscape of lncRNA isoforms in MM but also paves the way for novel RNA-targeted therapeutic strategies poised to transform cancer treatment.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal