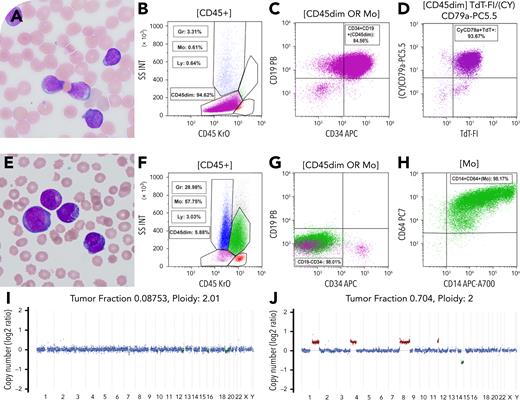
A newborn male presented with leukocytosis (white blood cells 139.6 × 109/L, hemoglobin 9.1 g/dL, platelets 70 × 109/L) and 51% circulating blasts. Morphology (panel A: Wright-Giemsa stain, 100× lens objective), flow cytometry (panels B-D), and fluorescence in situ hybridization confirmed congenital B-cell acute lymphoblastic leukemia (B-ALL) with KMT2A rearrangement. The patient received induction with an AALL15P1 regimen, followed by blinatumomab and consolidation chemotherapy. Central nervous system relapse occurred on day 90, with transient improvement on an ALL-MLL-10 regimen. By day 161, he suddenly developed recurrent leukocytosis (white blood cells 155.2 × 109/L). Morphology and immunophenotyping revealed transformation to acute myeloid leukemia (AML) with monocytic differentiation (panel E: Wright-Giemsa stain, 100× lens objective; and panels F-H). Whole-genome sequencing performed on baseline and transformation samples showed shared AFF1::KMT2A fusion and a KRAS p.G12D mutation. The variant allele frequency of KRAS increased from 44.3% (B-ALL) to 75.5% (AML), with concurrent copy-neutral loss of heterozygosity at 12p encompassing KRAS. Although the B-ALL genome lacked copy number variants (CNVs) (panel I), the AML genome exhibited additional CNVs, including gains of 1q, 4q, 8q, and 11q, and loss of 14q (panel J).
Despite escalation to intensive care, the patient died after transformation to AML. This case highlights the poor prognosis seen in blinatumomab-treated B-ALL with persistent KMT2A fusion and lineage switch to AML. Compared with conventional methods, serial genomic profiling by whole-genome sequencing may be more sensitive for detecting emerging genomic complexity (CNVs), and clonal evolution, in the setting of targeted therapy.
For additional images, visit the ASH Image Bank, a reference and teaching tool that is continually updated with new atlas and case study images. For more information, visit https://imagebank.hematology.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal