Abstract
In multiple myeloma, sequence studies of VH genes used to encode clonal Ig in neoplastic plasma cells have shown a common pattern of extensive somatic hypermutation. A further consistent feature of these VH sequences is a complete lack of intraclonal variation. These findings indicate that the malignant cell arises at a mature, postfollicular stage of B-cell development. However, only a minority of cases have a distribution of somatic mutations in VH consistent with a prior role for antigen in selecting the B cell of origin. To complement these studies, and to take further the investigation of a role for antigen in the clonal history of myeloma, we have investigated tumor-derived VL sequences from bone marrows of 15 patients. All sequences (9Vκ and 6Vλ) were potentially functional and 5 of 15 had evidence for N-region additions. All had undergone extensive somatic hypermutation, and showed no intraclonal variation. In 4 of 15 cases, the distribution of mutations revealed a significant (P < .05) clustering of replacement mutations in the CDR sequences, indicating a role for VL in selection by antigen. Comparison with the VH sequences used by the same tumor cells showed that, if significant clustering was present, it was in either VH or VL, but not both. Altogether, 10 of 15 V-regions showed evidence for antigen selection, suggesting that the B cell of origin has behaved as a normal germinal center B cell. Deductions concerning a role for antigen selection may require both VH and VL sequences for validation.
MULTIPLE MYELOMA is a malignant tumor involving plasma cells. Neoplastic cells are found in the bone marrow (BM), and typically secrete a monoclonal Ig of IgG or IgA isotype. Although the major identifiable tumor population consists of plasma cells, there has been a great deal of debate concerning the nature of the malignant cell, with some early indications that this may be a less mature B cell capable of “feeding” the plasma cell compartment.1,2 The advent of genetic technology aimed at Ig genes has allowed a more incisive investigation of the characteristics of myeloma clones. In fact, there have now been reports of a total of more than 50 sequences of VH genes used by tumor cells from patients' BM biopsies, and these have revealed common features.3-5
One conclusion is that usage of VH genes from the available repertoire appears to reflect no striking bias, with predominance of the large VH3 family in line with serological analysis of myeloma proteins.6 However, at the level of individual VH genes there may be some asymmetry in usage. For example, one gene, V4-34 , commonly used by normal B cells, and mandatory for encoding IgM autoanti–red blood cell antibodies of I/i specificity in patients with cold agglutinin disease,7 has so far not been found to be used by tumor cells in myeloma.5,8 In all cases of myeloma, the VH genes have been found to be somatically hypermutated.3-5 A further common feature is the lack of intraclonal variation in sequence, a finding that contrasts with the heterogeneity found in B-cell tumors of the germinal center.9,10 This leads to the conclusion that the final tumorigenic event in myeloma has occurred at a postfollicular stage, when the cell is no longer influenced by the somatic hypermutation mechanism.11 It argues against the concept that there is a “feeder” B cell, unless that cell has escaped the mutator before isotype switching.
However, IgM+ B cells with VH sequences indicating a clonal relationship with the neoplastic plasma cells have been detected in some cases of myeloma.12,13 Although there is some controversy about their frequency,14 it appears that such cells do exist and presumably continue to proliferate. There is uncertainty as to their contribution to malignancy, and it is possible that these cells have undergone some, but not all, of the events leading to malignant behavior.4
A further problem in understanding development of myeloma lies in the role of antigen in selecting the VH sequences of the tumor cell. If the cell of origin has been through somatic hypermutation, and antigen selection, before neoplastic transformation, this experience should be reflected in the V-gene sequences. For VH regions of antibody molecules, it is known that recognition of antigen can involve several sites, with CDR3 having a major influence.15 However, replacement mutations in CDR1 and CDR2 have a significant role in affinity maturation.11,16 For B-cell tumors, where the putative antigen is generally unknown, it is difficult to estimate involvement of CDR3 in recognition. In contrast, the possible clustering of replacement mutations in CDR1 and CDR2 which could be involved in affinity maturation can be analyzed. Rules to assess the significance of apparent clustering of replacement mutations compared with silent mutations have been developed.17 When these rules were applied to the large panel of VH sequences from myeloma cells, only a minority of cases (10 of 52) showed statistically significant clustering in CDRs.5 However, the antigen-binding site is known to involve both VH and VL ,18 and we have investigated VL sequences from a group of 15 patients both to extend our knowledge of Vκ and Vλ gene usage in myeloma, and to assess the role of VL in the selection of the cell of origin by antigen.
MATERIALS AND METHODS
Patients and cell preparation.Heparinized BM aspirates from unselected patients with multiple myeloma at different stages of disease from the Hematology (UK) or Immunology (Germany) clinics were taken for investigation. All patient material was obtained with consent, and with permission from local Ethical Committees. Clinical and laboratory features are shown in Table 1. All patients had an identifiable monoclonal Ig in serum (13 IgG, 2 IgA) of the same light chain type (9κ, 6λ) as the major plasma cell population. Mononuclear cells (MNC) were isolated by centrifugation on Ficoll-Hypaque (Pharmacia, Uppsala, Sweden), and plasma cell involvement was assessed by direct immunofluorescent staining for surface CD38 and cytoplasmic κ or λ light chains using the FACS-SCAN (Becton Dickinson, CA).19 In some cases, cytocentrifuged MNC preparations were stained with the same reagents, and assessed by fluorescent microscopy.
Characteristics of Myeloma Patients
| Patient . | Stage* . | Status . | Monoclonal Ig . | % Plasma Cells in BM . | |
|---|---|---|---|---|---|
| . | . | . | Class . | Level (g/L) . | . |
| 1 | IIIA | Relapsed | IgGκ | 24.0 | 11.0 |
| progressive | |||||
| 2 | IIA | Stable | IgGκ | 18.0 | 21.0 |
| 3 | II | Progressive | IgGκ | 43.0 | 5.0 |
| 4 | II | Stable | IgGκ | 28.0 | 30.0 |
| 5 | III | Progressive | IgGκ | 15.0 | 18.0 |
| 6 | IIA | Presentation | IgGκ | 59.0 | 14.0 |
| 7 | IIIA | Progressive | IgGκ | 71.0 | 30.0 |
| 8 | IB | Presentation | IgAκ | 40.7 | 60.0 |
| 9 | III | Progressive | IgGκ | 48.0 | 30.0 |
| 10 | IIA | Progressive | IgGλ | 20.0 | 19.0 |
| 11 | I | Stable | IgGλ | 16.0 | 4.0 |
| 12 | III | Progressive | IgGλ | 60.0 | 5.0 |
| 13 | III | Progressive | IgGλ | 21.0 | 10.0 |
| 14 | I | Progressive | IgAλ | 3.0 | 1.8 |
| 15 | IIIA | Presentation | IgGλ | 79.0 | 50.0 |
| Patient . | Stage* . | Status . | Monoclonal Ig . | % Plasma Cells in BM . | |
|---|---|---|---|---|---|
| . | . | . | Class . | Level (g/L) . | . |
| 1 | IIIA | Relapsed | IgGκ | 24.0 | 11.0 |
| progressive | |||||
| 2 | IIA | Stable | IgGκ | 18.0 | 21.0 |
| 3 | II | Progressive | IgGκ | 43.0 | 5.0 |
| 4 | II | Stable | IgGκ | 28.0 | 30.0 |
| 5 | III | Progressive | IgGκ | 15.0 | 18.0 |
| 6 | IIA | Presentation | IgGκ | 59.0 | 14.0 |
| 7 | IIIA | Progressive | IgGκ | 71.0 | 30.0 |
| 8 | IB | Presentation | IgAκ | 40.7 | 60.0 |
| 9 | III | Progressive | IgGκ | 48.0 | 30.0 |
| 10 | IIA | Progressive | IgGλ | 20.0 | 19.0 |
| 11 | I | Stable | IgGλ | 16.0 | 4.0 |
| 12 | III | Progressive | IgGλ | 60.0 | 5.0 |
| 13 | III | Progressive | IgGλ | 21.0 | 10.0 |
| 14 | I | Progressive | IgAλ | 3.0 | 1.8 |
| 15 | IIIA | Presentation | IgGλ | 79.0 | 50.0 |
Durie and Salmon staging.
Analysis of VL genes.For preparation of cDNA, total RNA (2 to 10 μg) was isolated from the MNC fraction (1 to 6 × 106 cells) using RNAzol B (Cinna Biotecx Labs Inc, Houston, TX). Reverse transcription was carried out with an oligo dT primer, using a first-strand cDNA synthesis kit (Pharmacia). A sample of the cDNA (1/3 to 1/5) was then amplified by polymerase chain reaction (PCR) using a mixture of 5′ oligonucleotide FWR1 primers specific for the expressed Vκ or Vλ families together with a mixture of downstream 3′ primers specific for Jκ or Jλ genes as appropriate (Table 2). Amplification conditions were as described,4,20 except that annealing temperature was 65°C for 1 minute. Amplified products were cloned and sequenced as described4; alignment was made to current EMBL/GenBank and V-BASE sequence directories21 using MacVector 4.0 sequence analysis software (International Biotechnologies Inc, New Haven, CT). At least two independent PCR amplifications were performed from each sample.
Oligonucleotide VL PCR Primers
| Primer . | Location . | Orientation . | Sequence (5′-3′) . |
|---|---|---|---|
| Vκ1&4 | FR1 | Sense | GACATCSWGATGACCCAGTCTCC |
| Vκ2&6 | FR1 | Sense | GAWRTTGTGMTGACTCAGTCTCC |
| Vκ3 | FR1 | Sense | GAAATTGTGTTGACGCAGTCTCC |
| Vκ5 | FR1 | Sense | GAAACGACACTCACGCAGTCTCC |
| Jκ1-4 | FR4 | Anti-sense | ACGTTTGATHTCCACYTTGGTCCC |
| Jκ5 | FR4 | Anti-sense | ACGTTTAATCTCCAGTCGTGTCCC |
| Vλ1 | FR1 | Sense | CAGTCTGTSBTGACKCAGCCRCCY |
| Vλ2 | FR1 | Sense | CAGTCTGCCCTGACTCAGCCTSSYT |
| Vλ3 | FR1 | Sense | TCYTMTGWGCTGACTCAGSMM |
| Vλ7&8 | FR1 | Sense | CAGRCTGTGGTGACYCAGGAGCCMTC |
| Vλ9 | FR1 | Sense | CAGCCTGTGCTGACTCAGCCACCTTC |
| JλC | FR4 | Anti-sense | ACCKAGGACGGTSASCTKGGTSCC |
| Primer . | Location . | Orientation . | Sequence (5′-3′) . |
|---|---|---|---|
| Vκ1&4 | FR1 | Sense | GACATCSWGATGACCCAGTCTCC |
| Vκ2&6 | FR1 | Sense | GAWRTTGTGMTGACTCAGTCTCC |
| Vκ3 | FR1 | Sense | GAAATTGTGTTGACGCAGTCTCC |
| Vκ5 | FR1 | Sense | GAAACGACACTCACGCAGTCTCC |
| Jκ1-4 | FR4 | Anti-sense | ACGTTTGATHTCCACYTTGGTCCC |
| Jκ5 | FR4 | Anti-sense | ACGTTTAATCTCCAGTCGTGTCCC |
| Vλ1 | FR1 | Sense | CAGTCTGTSBTGACKCAGCCRCCY |
| Vλ2 | FR1 | Sense | CAGTCTGCCCTGACTCAGCCTSSYT |
| Vλ3 | FR1 | Sense | TCYTMTGWGCTGACTCAGSMM |
| Vλ7&8 | FR1 | Sense | CAGRCTGTGGTGACYCAGGAGCCMTC |
| Vλ9 | FR1 | Sense | CAGCCTGTGCTGACTCAGCCACCTTC |
| JλC | FR4 | Anti-sense | ACCKAGGACGGTSASCTKGGTSCC |
RESULTS
VL gene usage by tumor cells.The VL genes used by the tumor cells were identified as repeated identical VL-JL sequences obtained after cloning of PCR products.4 Remaining sequences, presumably from normal B cells in the aspirates,20 were different from each other. Repeated sequences were seen in all cases, at variable frequency, as indicated in Tables 3 and 4 for Vκ and Vλ, respectively. The profile of Vκ genes used by the 9 κ-positive tumors indicates that 5 of 9 use Vκ1 and 4 of 9 VκIII frequencies in line with normal B cells.22,23 Among the Vκ1 group, 4 of 5 use the O8/18 gene, which is commonly rearranged in B-cell tumors,24 and 3 of 4 of the VκIII group use the A27 gene, found to be used frequently in chronic lymphocytic leukemia.25 The Vλ genes (Table 4) used three different families. There appeared to be no preferential use of particular JL genes for either light chain type.
Analysis of Vκ Genes From Myeloma Patients
| Patient No. . | Ig Light Chain . | VL Family . | GL Donor . | % Homology . | R/S Mutations . | JL . | Tumor-Derived Sequences/Clones Sequenced . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | FWR . | CDR . | . | . |
| 1 | κ | Vκ I | O8/18 | 93.2 | 5/8 | 4/1 | Jκ 5 | 8/11 |
| 2 | κ | Vκ I | O8/18 | 93.8 | 6/2 | 5/3 | Jκ 2 | 8/12 |
| 3 | κ | Vκ I | O8/18 | 94.6 | 4/5 | 3/2 | Jκ 4 | 9/11 |
| 4 | κ | Vκ I | O8/18 | 94.7 | 3/5 | 2/4 | Jκ 4 | 7/9 |
| 5 | κ | Vκ I | A30 | 95.0 | 5/3 | 5/0 | Jκ 1 | 8/11 |
| 6 | κ | Vκ III | A27 | 93.6 | 6/2 | 6/4 | Jκ 1 | 8/8 |
| 7 | κ | Vκ III | A27 | 95.0 | 4/1 | 6/2 | Jκ 4 | 10/12 |
| 8 | κ | Vκ III | A27 | 93.9 | 4/1 | 11/0 | Jκ 2 | 12/12 |
| 9 | κ | Vκ III | L6 | 98.5 | 0/0 | 3/1 | Jκ 3 | 6/11 |
| Patient No. . | Ig Light Chain . | VL Family . | GL Donor . | % Homology . | R/S Mutations . | JL . | Tumor-Derived Sequences/Clones Sequenced . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | FWR . | CDR . | . | . |
| 1 | κ | Vκ I | O8/18 | 93.2 | 5/8 | 4/1 | Jκ 5 | 8/11 |
| 2 | κ | Vκ I | O8/18 | 93.8 | 6/2 | 5/3 | Jκ 2 | 8/12 |
| 3 | κ | Vκ I | O8/18 | 94.6 | 4/5 | 3/2 | Jκ 4 | 9/11 |
| 4 | κ | Vκ I | O8/18 | 94.7 | 3/5 | 2/4 | Jκ 4 | 7/9 |
| 5 | κ | Vκ I | A30 | 95.0 | 5/3 | 5/0 | Jκ 1 | 8/11 |
| 6 | κ | Vκ III | A27 | 93.6 | 6/2 | 6/4 | Jκ 1 | 8/8 |
| 7 | κ | Vκ III | A27 | 95.0 | 4/1 | 6/2 | Jκ 4 | 10/12 |
| 8 | κ | Vκ III | A27 | 93.9 | 4/1 | 11/0 | Jκ 2 | 12/12 |
| 9 | κ | Vκ III | L6 | 98.5 | 0/0 | 3/1 | Jκ 3 | 6/11 |
Analysis of Vλ Genes From Myeloma Patients
| Patient No. . | Ig Light Chain . | VL Family . | GL Donor . | % Homology . | R/S Mutations . | JL . | Tumor-Derived Sequences/Clones Sequenced . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | FWR . | CDR . | . | . |
| 10 | λ | Vλ I | DPL2 | 95.6 | 3/3 | 4/2 | Jλ 7 | 12/12 |
| 11 | λ | Vλ I | DPL3 | 91.0 | 3/6 | 9/6 | Jλ 1 | 4/10 |
| 12 | λ | Vλ III | IGLV3S2 | 96.5 | 0/2 | 6/1 | Jλ 7 | 8/8 |
| 13 | λ | Vλ II | DPL11 | 95.9 | 2/4 | 5/0 | Jλ 2 | 6/9 |
| 14 | λ | Vλ II | HSLV2046 | 94.4 | 4/3 | 6/2 | Jλ 2 | 9/11 |
| 15 | λ | Vλ III | DPL23 | 92.9 | 3/6 | 7/2 | Jλ 2 | 8/8 |
| Patient No. . | Ig Light Chain . | VL Family . | GL Donor . | % Homology . | R/S Mutations . | JL . | Tumor-Derived Sequences/Clones Sequenced . | |
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | FWR . | CDR . | . | . |
| 10 | λ | Vλ I | DPL2 | 95.6 | 3/3 | 4/2 | Jλ 7 | 12/12 |
| 11 | λ | Vλ I | DPL3 | 91.0 | 3/6 | 9/6 | Jλ 1 | 4/10 |
| 12 | λ | Vλ III | IGLV3S2 | 96.5 | 0/2 | 6/1 | Jλ 7 | 8/8 |
| 13 | λ | Vλ II | DPL11 | 95.9 | 2/4 | 5/0 | Jλ 2 | 6/9 |
| 14 | λ | Vλ II | HSLV2046 | 94.4 | 4/3 | 6/2 | Jλ 2 | 9/11 |
| 15 | λ | Vλ III | DPL23 | 92.9 | 3/6 | 7/2 | Jλ 2 | 8/8 |
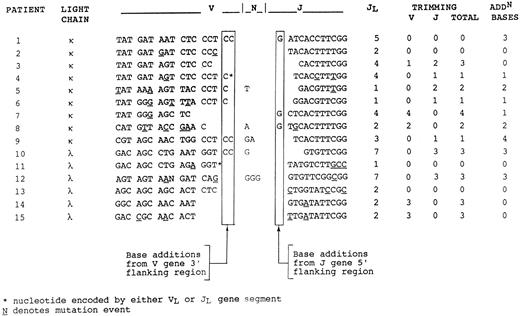
V-J joining region.Clonality of tumor-derived sequences was confirmed by analysis of the V-J junction (Fig 1), which showed intraclonal identity. In 9 of 15 sequences, there were base additions at the junction which were not encoded by the V or J genes. In some cases, these appeared to be derived from flanking regions of the genes, and could therefore be accounted for by imprecision at the joint. In 5 of 15, there were additional nucleotides which may represent N-region additions, contributed by TdT activity. In a majority (11 of 15) of cases, nucleotides had been lost by trimming from either V or J genes.
Nucleotide sequences of the V-J junctional regions. Junctional regions are identical in the repeated sequences from each individual case (Tables 3 and 4). Base additions from flanking regions and losses by trimming are indicated. Remaining bases are presumed to have arisen via N-region addition.
Nucleotide sequences of the V-J junctional regions. Junctional regions are identical in the repeated sequences from each individual case (Tables 3 and 4). Base additions from flanking regions and losses by trimming are indicated. Remaining bases are presumed to have arisen via N-region addition.
Somatic mutation.Nucleotide sequences of all VL genes have been submitted to the EMBL/GenBank database (accession numbers Z70253-255; Z70258-261; Z70263-264; Z75558; X98894-898). To assess the degree of somatic hypermutation, comparison with the closest counterparts in the database of germline sequences has been made. This does not take into account any polymorphisms in VL , but these are known to be insignificant in Vκ.26 Less information is available for Vλ genes, but again suggests only limited polymorphic variation.27 The VL sequences obtained deviated significantly from the closest germline genes in the database (Tables 3 and 4), with a mean percent mutation of 5.3 for Vκ and 6.2 for Vλ. There was evidence for block mutations, involving two or more adjacent nucleotides, in both light-chain types. For Vκ, the high incidence of block mutations (6 of 9 sequences [67%]) compares with the reported figure of ∼50%.28 The numerical distribution of the mutations in FWRs and CDRs, and the ratio of replacement to silent mutations are shown in Tables 3 and 4. Deduced amino acid sequences are shown in Figs 2 and 3. In all cases, somatic mutations were identified in the VL sequences, with 9 of 15 having additional identifiable mutations in JL , even though events at the 3′ end of JL are obscured by the primer sites. Several sequences derived from the same VL family member were available for the Vκ genes O8/18 and A27. Comparison of these showed no evidence for common sites or “hot-spots” of mutational activity. Analysis of the distribution of somatic mutations in each sequence (Table 5) has been carried out by the method of Chang and Casali.17 In this method, each VL or VH gene sequence is assessed codon by codon for significance of deviation from germline sequence. A modification of the binomial distribution model is then used to calculate whether the probability (P in Tables 5 and 6) of an excess (in CDRs) or scarcity (in FWRs) of replacement mutations resulted by chance alone.17 For the FWRs, there were generally fewer replacement (R) mutations than expected due to chance, with significant (P < .05) conservation of sequence in 10 of 15 sequences, a feature commonly seen for VH .14 For the CDRs, there were more R mutations than expected in 14 of 15 sequences, with significant (P < .05) clustering indicative of antigen selection in 4 of 15 (3Vκ and 1Vλ).
Deduced amino acid sequences of the Vκ regions of the tumor-derived clones from patients with myeloma. Comparisons are made with the closest germline Vκ genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in Jκ are underlined.
Deduced amino acid sequences of the Vκ regions of the tumor-derived clones from patients with myeloma. Comparisons are made with the closest germline Vκ genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in Jκ are underlined.
Deduced amino acid sequences of the Vλ regions of the tumor-derived clones from patients with myeloma. Comparisons are made with the closest germline Vλ genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in Jλ are underlined.
Deduced amino acid sequences of the Vλ regions of the tumor-derived clones from patients with myeloma. Comparisons are made with the closest germline Vλ genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in Jλ are underlined.
R and S Mutations in Myeloma VL Genes
| Patient . | Ig Class . | Germline Gene . | R:S (CDR)obs R:S (FWR)obs . | R (CDR)exp R (FWR)exp . | P (CDR)* P (FWR) . |
|---|---|---|---|---|---|
| 1 | IgGκ | 08/18 | 4.00 (4:1) | 4 | .22 |
| 0.63 (5:8) | 10 | .02 | |||
| 2 | IgGκ | 08/18 | 1.70 (5:3) | 4 | .14 |
| 3.00 (6:2) | 9 | .09 | |||
| 3 | IgGκ | 08/18 | 1.50 (3:2) | 3 | .26 |
| 0.80 (4:5) | 8 | .03 | |||
| 4 | IgGκ | 08/18 | 0.50 (2:4) | 3 | .20 |
| 0.60 (3:5) | 8 | .01 | |||
| 5 | IgGκ | A30 | 5.00 (5:0) | 3 | .09 |
| 1.00 (4:4) | 7 | .19 | |||
| 6 | IgGκ | A27 | 1.50 (6:4) | 4 | .12 |
| 3.00 (6:2) | 10 | .16 | |||
| 7 | IgGκ | A27 | 3.00 (6:2) | 3 | .03 |
| 4.00 (4:1) | 7 | .06 | |||
| 8 | IgAκ | A27 | ∞ (11.0) | 3 | .0001 |
| 4.00 (4:1) | 8 | .02 | |||
| 9 | IgGκ | L6 | ∞ (3:0) | 1 | .04 |
| 0.00 (0:1) | 2 | .00 | |||
| 10 | IgGλ | DPL2 | 2.00 (4:2) | 3 | .20 |
| 1.00 (3:3) | 6 | .06 | |||
| 11 | IgGλ | DPL3 | 1.50 (9:6) | 6 | .08 |
| 0.50 (3:6) | 12 | .0002 | |||
| 12 | IgGλ | IGLV3S2 | 6.00 (6:1) | 2 | .006 |
| 0.00 (0:2) | 0 | .00 | |||
| 13 | IgGλ | DPL11 | 5.00 (5:0) | 3 | .08 |
| 0.50 (2:4) | 6 | .03 | |||
| 14 | IgAλ | HSLV2046 | 3.00 (6:2) | 4 | .09 |
| 1.33 (4:3) | 8 | .04 | |||
| 15 | IgGλ | DPL23 | 3.50 (7:2) | 4 | .14 |
| 0.50 (3:6) | 10 | .0012 |
| Patient . | Ig Class . | Germline Gene . | R:S (CDR)obs R:S (FWR)obs . | R (CDR)exp R (FWR)exp . | P (CDR)* P (FWR) . |
|---|---|---|---|---|---|
| 1 | IgGκ | 08/18 | 4.00 (4:1) | 4 | .22 |
| 0.63 (5:8) | 10 | .02 | |||
| 2 | IgGκ | 08/18 | 1.70 (5:3) | 4 | .14 |
| 3.00 (6:2) | 9 | .09 | |||
| 3 | IgGκ | 08/18 | 1.50 (3:2) | 3 | .26 |
| 0.80 (4:5) | 8 | .03 | |||
| 4 | IgGκ | 08/18 | 0.50 (2:4) | 3 | .20 |
| 0.60 (3:5) | 8 | .01 | |||
| 5 | IgGκ | A30 | 5.00 (5:0) | 3 | .09 |
| 1.00 (4:4) | 7 | .19 | |||
| 6 | IgGκ | A27 | 1.50 (6:4) | 4 | .12 |
| 3.00 (6:2) | 10 | .16 | |||
| 7 | IgGκ | A27 | 3.00 (6:2) | 3 | .03 |
| 4.00 (4:1) | 7 | .06 | |||
| 8 | IgAκ | A27 | ∞ (11.0) | 3 | .0001 |
| 4.00 (4:1) | 8 | .02 | |||
| 9 | IgGκ | L6 | ∞ (3:0) | 1 | .04 |
| 0.00 (0:1) | 2 | .00 | |||
| 10 | IgGλ | DPL2 | 2.00 (4:2) | 3 | .20 |
| 1.00 (3:3) | 6 | .06 | |||
| 11 | IgGλ | DPL3 | 1.50 (9:6) | 6 | .08 |
| 0.50 (3:6) | 12 | .0002 | |||
| 12 | IgGλ | IGLV3S2 | 6.00 (6:1) | 2 | .006 |
| 0.00 (0:2) | 0 | .00 | |||
| 13 | IgGλ | DPL11 | 5.00 (5:0) | 3 | .08 |
| 0.50 (2:4) | 6 | .03 | |||
| 14 | IgAλ | HSLV2046 | 3.00 (6:2) | 4 | .09 |
| 1.33 (4:3) | 8 | .04 | |||
| 15 | IgGλ | DPL23 | 3.50 (7:2) | 4 | .14 |
| 0.50 (3:6) | 10 | .0012 |
Probability calculations according to Chang and Casali.17
Comparison of Antigen-Driven R Mutations Locating to Myeloma VH or VL Genes
| Patient . | P (CDR)6-150 . | |
|---|---|---|
| . | VH . | VL . |
| 1 | .10 | .22 |
| 2 | .03 | .14 |
| 3 | .06 | .25 |
| 4 | .18 | .20 |
| 5 | .11 | .09 |
| 6 | .0009 | .12 |
| 7 | .13 | .03 |
| 8 | .21 | .0001 |
| 9 | .25 | .04 |
| 10 | .0029 | .20 |
| 11 | .0098 | .08 |
| 12 | .10 | .006 |
| 13 | .049 | .08 |
| 14 | .18 | .09 |
| 15 | .04 | .14 |
| Patient . | P (CDR)6-150 . | |
|---|---|---|
| . | VH . | VL . |
| 1 | .10 | .22 |
| 2 | .03 | .14 |
| 3 | .06 | .25 |
| 4 | .18 | .20 |
| 5 | .11 | .09 |
| 6 | .0009 | .12 |
| 7 | .13 | .03 |
| 8 | .21 | .0001 |
| 9 | .25 | .04 |
| 10 | .0029 | .20 |
| 11 | .0098 | .08 |
| 12 | .10 | .006 |
| 13 | .049 | .08 |
| 14 | .18 | .09 |
| 15 | .04 | .14 |
Probability calculations according to Chang and Casali.17
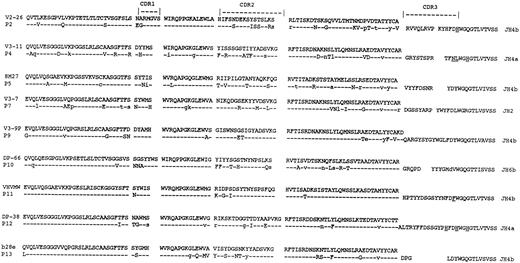
Comparison with VH genes.For 6 cases (patients 1, 3, 6, 8, 14, and 15), tumor-derived VH gene sequences were known.4,29 VH sequences from the remaining 9 patients were obtained as described4 and deduced amino acid sequences are shown in Fig 4. Nucleotide sequences have been submitted to EMBL/GenBank database (accession numbers: Z70256-257; Z75556-5557; X98899-99003). Although the closest germline gene has been obtained from the database, rather than from the patients' DNA, it appears that in general polymorphism in VH is not sufficient to require this approach.21 In fact, where we4 and others5 have analyzed the patients' germline VH genes, the sequence has been found in the majority of cases to correspond exactly to that obtained from the database. However, in 1 of 9 cases of myeloma, a 2-bp difference from the published sequence of a VII-5 germline gene was found also in the patient's germline sequence, indicating that this was probably caused by a polymorphism.5 The distribution of somatic mutations in VH of 6 of 15 of these cases indicated a significant clustering in CDRs (Table 6). Comparison of patterns in VH with those in VL (Table 6) showed that clustering in CDRs of VH was not paralleled by clustering in CDRs of VL . In addition, the clustering in VL observed in 4 of 15 cases was not paralleled by clustering in VH . Therefore, from the 10 cases where clustering was evident, it was localized in either VH or VL , but not both. However, in 5 of 15 cases, there was no significant clustering in either VH or VL .
Deduced amino acid sequences of the VH regions of tumor cells from patients with myeloma. Comparisons are made with the closest germline VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined. Patient identification numbers are indicated. VH sequences from other patients are published.4 37
Deduced amino acid sequences of the VH regions of tumor cells from patients with myeloma. Comparisons are made with the closest germline VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined. Patient identification numbers are indicated. VH sequences from other patients are published.4 37
DISCUSSION
Analysis of V-genes used by neoplastic B cells is extending our understanding of the origin and progression of B-cell tumors. Now that a complete map of the VH gene germline repertoire is available,21,30 it is possible to compare a VH sequence from a tumor cell to the germline gene of origin with confidence. Although some nucleotide changes may reflect polymorphic variation, particularly for certain VH3 genes,31 it can be assumed that the majority of deviations from germline sequence in VH genes of a B cell represent somatic mutations.21 In some cases this has been proved by comparing the tumor-derived sequence with the germline counterpart in the patient.4,5 Accumulation of such mutations indicate that the cell of origin has been exposed to the hypermutation mechanism in the germinal center.11,18,32 Heterogeneity of mutations within the tumor clone indicates that the tumor cell is still under the influence of the mutation mechanism, subsequent to neoplastic transformation.9,10 Finally, concentration of replacement mutations in CDRs can suggest a role for antigen in selection of the B cell.17 33
In the case of multiple myeloma, VH gene analyses from several laboratories have shown that the malignant cell has undergone extensive somatic hypermutation.3-5 There is further wide agreement that there is no intraclonal heterogeneity among the tumor cell population,3-5 and there is evidence that the VH sequence is stable from diagnosis through plateau phase.34 These findings strongly suggest that the malignant cell has exited from the germinal center, and is no longer susceptible to the mutation mechanism.4 5
The germline repertoire of VL genes has also been mapped,27,35,36 but there have been fewer studies of usage in B-cell tumors. In myeloma, using DNA as a source, 7 Vκ sequences were obtained from 29 cases, with 4 of 7 potentially functional.37 Sequences were somatically mutated, with a hint of antigen selection from R:S ratios.37 A second study investigated Vκ-gene usage in 3 cases of myeloma.24 Together, these studies showed that 3 of 7 functional genes were derived from the O8/18 gene,24,37 and we have confirmed this incidence (4 of 9 cases). Although the Vκ1 family is often used by normal B cells,22,23 the level of usage of the O8/18 gene appears high in myeloma. However, frequency of this gene in other B-cell tumors has also been reported to be high, and it is not yet clear if there is a difference among the tumor categories.24
The current results have focused on functional VL genes, obtained from RNA. Identification of repeated sequences in the cloned PCR product supports the derivation from tumor cells, which can be a problem otherwise. We have analyzed the pattern of both Vκ and Vλ sequences. These confirm the high level of somatic hypermutation, with the level of 5.8% mutation for VL being comparable with that of 8.2% for VH .5 There is also a lack of intraclonal heterogeneity in VL of all patients, again confirming findings in VH , and supporting the concept that the final event in malignant transformation has occurred at a postfollicular stage.3,4,14 In contrast, the VH genes in the benign counterpart of myeloma (monoclonal gammopathy of undetermined significance or MGUS), showed intraclonal heterogeneity in 3 of 7 cases.29 This could indicate that the clonal plasma cell in MGUS is less mature, and may have undergone some, but not all the events leading to malignant behavior.29
If the final neoplastic event is late in maturation of the B cell, it might be expected that the myeloma precursor cell will have been subjected to the same processes of development as a normal B cell. Even if there is an IgM+ clonal precursor, which has undergone some neoplastic event, the few cases available for analysis have indicated that it has a homogeneous VH gene sequence identical to the isotype-switched plasma cell.12,13,38 This would argue that neoplastic transformation in myeloma begins in a mature B cell immediately before isotype-switch. Because a B cell would have reached this point following antigen selection, the imprint of this procedure should remain as a clustering of mutations in CDRs of V-gene sequences.11,17,33 In fact, analysis of the stable sequences in myeloma should be particularly useful, because the selected sequence will not be obliterated by continuing posttransformation mutations. However, analysis of VH sequences in myeloma has given mixed results, with only 21% of the tumor-derived sequences from a large series showing significant clustering in CDRs.5,14 This leaves open the question of the clonal history of the tumor cells in the remaining 79% of the cases. Because VL sequence is also known to be involved in recognition of antigen,18 deductions from V-gene sequences that relate to a role for antigen in selection should be strengthened by including analysis of VL .
In this study, significant clustering of mutations in CDRs of VL was seen in 4 of 15 sequences. In contrast, a preliminary report of a study of Vκ sequences in 9 cases of myeloma has indicated that clustering of replacement mutations in CDRs occurred in all cases, but more details of the analysis are required.39 In our cases, comparison with VH sequence in the same cell showed that clustering was in either VH or VL , but not both. This suggests that a role for antigen might be more common than estimated from VH alone, reaching 67% in our study. There have been insufficient studies of normal human B cells to know if this is a typical finding. Investigation of the classical murine response to the hapten phenyl oxazolone has shown that affinity maturation is accompanied by somatic mutations in the CDRs of both VH and Vκ.40 This is likely to be the case in human antibodies, and might suggest that a role for antigen is more common than estimated from VH alone. For myeloma, the findings support the idea that the cell of origin has undergone the process of conventional antigen selection. However, there remains a minority of sequences which appear to have no clustering in either V-region. Even this pattern does not rule out a role for antigen selection, because optimal binding may occur via CDR3. Clearly we need more information on how normal human B cells generate antibody, but this study would suggest that deductions concerning a role for antigen in the clonal history of neoplastic B cells should take into account mutational events in both VH and VL .
ACKNOWLEDGMENT
We thank Dr D.G. Oscier for providing patient material and for helpful comments.
Supported by the Leukaemia Research Fund, UK, the European Myeloma Research Network (Biomed BMH1-CT93-1407), and the Dr Hiltrud Pulst Myeloma Foundation, Hannover, Germany.
Address reprint requests to Freda K. Stevenson, DPhil, Molecular Immunology Group, Tenovus Laboratory, Southampton University Hospitals, Tremona Rd, Southampton SO16 6YD, UK.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal