Abstract
Hematopoietic stem and progenitor cell populations were obtained by fluorescence activated cell sorting of murine bone marrow (BM) cells into Rhodamine-123lo lineage−Ly6A/E+ c-kit+ (primitive stem cells highly enriched for long-term BM repopulating activity), Rhodamine-123med/hi lineage− Ly6A/E+ c-kit+ (mature stem cells highly enriched for short-term BM repopulating activity and day 13 spleen colony-forming activity) and lineage− Ly6A/E− c-kit+ (enriched for in vitro colony forming cells) populations. Neither stem cell population responds to single cytokines in vitro and each requires the synergistic action of two or more cytokines for proliferation, whereas the progenitor cell population proliferates in response to single cytokines. Since each of these cell populations was sorted as c-kit+, they express receptors for stem cell factor. Cell populations were also analyzed by autoradiography for their ability to specifically bind iodinated cytokines and this revealed that both stem cell populations expressed receptors for interleukin-1α (IL-1α), IL-3, IL-6, and granulocyte colony-stimulating factor (G-CSF ), but lacked receptors for macrophage colony-stimulating factor (M-CSF ), granulocyte-macrophage colony stimulating factor (GM-CSF ), and leukemia inhibitory factor (LIF ). Cells within the progenitor cell population specifically bound IL-3, GM-CSF, G-CSF, IL-6, and IL-1α, whereas no receptors were detected for M-CSF and LIF. Within each cell population examined, heterogeneity was observed in the percentage of cells labeled and the number of receptors per cell. These results suggest that stem cell populations can be further subdivided according to their cytokine receptor profile and it will be of interest to determine if such subpopulations have distinctive functional properties.
THE HEMATOPOIETIC stem cells (HSC) and their immediate progeny, the committed progenitor cells, are able to be distinguished by their differing capacities for self-renewing proliferation and degree of multipotentiality. However, the cells of both populations are heterogeneous and a hierarchy of stem cells exists based on varying capacities for self-generation and proliferation. The most primitive HSCs are cells having the maximal proliferative, self-renewal, and bone marrow (BM) repopulating ability. The progeny of these cells sustain short-term BM repopulation as a result of their reduced capacity to self-renew, form spleen colonies in irradiated animals and blast colonies or large colonies containing multiple cell lineages in in vitro colony-forming assays.1
HSCs constitute only 0.01% of cells in the BM of adult mice. Fluorescence activated cell sorting (FACS) has allowed the isolation of highly enriched populations of HSCs according to the degree to which they incorporate the mitochondrial supravital dye Rhodamine-123 (Rh), express the surface markers c-kit (receptor for stem cell factor [SCF]) and Ly6A/E (Sca-1), and lack lineage markers specific for T, B, and myeloid cells (lin−).1-8
A variety of in vitro bioassays have been used to establish the responsiveness of HSCs to stimulation by various cytokines. However, interpretation of the data has been complicated by the possibility of indirect actions, and the observation that highly enriched populations of HSCs do not in general respond to single cytokines, but require the presence of two or more cytokines before initiating a mitogenic response,7-14 possibly through the activation of multiple signaling pathways.15-19
To overcome this problem and gain an insight into the cytokines that might have direct individual or synergistic actions on HSCs and progenitor cells, the expression of cytokine receptors on these cells was investigated using radiolabeled cytokines.
After binding of radiolabeled ligand by cells, Scatchard analysis20 provides information on mean receptor numbers per cell and the affinity of the interaction between the cytokine and its corresponding receptor. However, with heterogeneous populations of cells, the contribution of specific cell types to the binding reaction can only be determined by using either pre-enriched populations or by autoradiography of cell populations. These approaches are superior to studies investigating mRNA levels for receptors by Northern analysis, or the more sensitive polymerase chain reaction (PCR), because analysis of mRNA cannot establish the level of translation of a receptor product or its level of expression at the cell surface.
In the present study, murine HSCs were sorted into two populations; primitive cells (Rhlo lin− Ly6A/E+ c-kit+) that contributed to long-term BM repopulation of lethally irradiated animals, and a more mature HSC population (Rhhi lin− Ly6A/E+ c-kit+) enriched for short-term BM-repopulation activities. Using this protocol, it was possible to reproducibly obtain at least 20,000 to 30,000 HSCs of each type from the BM cells of 10 to 12 mice. A highly enriched population of committed progenitor cells was also obtained using the sorting parameters lin− Ly6A/E− c-kit+. Sufficent numbers of cells were obtained to permit cell autoradiography but not direct Scatchard analysis. However, parallel Scatchard analysis and autoradiography on unfractionated normal adult murine BM allowed a correlation to be established between receptor number and the number of silver grains observed over enriched HSC and progenitor cells.
MATERIALS AND METHODS
Animals.C57BL/Ka-Thy-1.1 (Thy-1.1, Ly-5.2) and C57BL/6-Ly-5.1-Pep3b (Thy-1.2, Ly-5.1) mouse strains at 6 to 12 weeks of age were used throughout the experiments. They were bred at The Walter and Eliza Hall Institute of Medical Research animal facility under specific pathogen-free conditions and were maintained on acidified water.
Preparation of FACS sorted cells.BM cells from 10 8- to 12-week-old C57BL/Ka-Thy-1.1, Ly-5.2 mice were sorted into the following populations; Rhlolin− Ly6A/E+ c-kit+ cells, Rhhilin− Ly6A/E+ c-kit+, and lin− Ly6A/E− c-kit+ according to our published protocol.1 All cells were maintained at 4°C throughout the experiment. After completion of cell sorting, the deflected cells were centrifuged at 340g for 5 minutes, resuspended, and counted using a hemocytometer.
Cytokines.All cytokines used had been purified to apparent homogeneity. Recombinant human interleukin-1α (rh-IL-1α) was produced in Escherichia coli and obtained from Immunex Corp (Seattle, WA). Recombinant murine interleukin-3 (rm-IL-3) was produced in E coli and was purchased from Preprotech (Rocky Hill, NJ). Recombinant murine interleukin-6 (rm-IL-6) produced in E coli was a gift from Dr R.J. Simpson (Ludwig Institute for Cancer Research, Melbourne, Australia). Recombinant rat stem cell factor (rr′-SCF ) was a gift from Dr K. Zsebo (Amgen Corp, Thousand Oaks, CA). Recombinant murine granulocyte-macrophage colony stimulating factor (rm-GM-CSF ) produced in E coli was a gift from Schering Plough (Kenilworth, NJ). Native mouse granulocyte-colony stimulating factor (nm-G-CSF ), recombinant murine macrophage-colony stimulating factor (rm-M-CSF ) produced in yeast, and recombinant murine and human leukemia inhibitory factor (rm-LIF and rh-LIF ) both produced in E coli, were purified in the Cancer Research Unit at The Walter and Eliza Hall Institute of Medical Research.
Radioiodination of cytokines.125I-human IL-1α was purchased from Amersham (Arlington Heights, IL) and had a specific activity of 2 × 105 cpm/ng. All other cytokines were radioiodinated using the modified iodine monochloride method.21 Briefly 1 to 5 μg of purified cytokine in 5 to 100 μL aqueous solution was radioiodinated with the addition of 1 mCi (3 μL) carrier free Na125I (New England Nuclear, North Ryde, NSW, Australia), and 40 μL 0.2 mol/L sodium phosphate buffer pH 7.2 containing 0.02% Tween-20 (vol/vol). While vortexing, 3 μL 0.2 mmol/L ICl in 2.0 mol/L NaCl (produced as described by Contreras, 1983)21 was added. After 30 seconds at room temperature, a further 6 μL of the ICl solution was added. The reaction mixture was chromatographed on a PD-10 column (Pharmacia, Uppsala, Sweden) previously equilibrated in 20 mmol/L sodium phosphate buffer pH 7.3 containing 0.15 mol/L NaCl, 0.02% Tween-20 (vol/vol), and 0.02% sodium azide (wt/vol) phosphate-buffered saline (PBS).
After radioiodination, proteins were separated from free 125I and other impurities by ion exchange chromatography. Fractions containing the radioiodinated protein were diluted 10-fold in 10 mmol/L citric acid containing 0.02% Tween-20 (vol/vol) and chromatographed on a 100 μL column of CM-Sepharose CL-6B (Pharmacia) equilibrated in the same buffer. The column was washed with 5 mL equilibration buffer and the radio-iodinated protein eluted with five 0.5 mL aliquots of Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum (FCS).
The bindable fraction and specific radioactivity of each 125I-cytokine were determined by the method of Calvo et al23 and were for SCF 0.3 and 3 × 104 cpm/ng; M-CSF 1.0 and 7.5 × 105 cpm/ng; IL-3 0.93 and 2.7 × 105 cpm/ng; GM-CSF 0.5 and 1.25 × 105 cpm/ng; G-CSF 1.0 and 2× 104 cpm/ng; IL-6 0.3 and 2.5 × 105 cpm/ng; LIF 1.0 and 4.5 × 104 cpm/ng and IL-1α not determined and 2 × 105 cpm/ng, respectively.
Binding of 125I-cytokines to cells.Aliquots of each of the sorted cell populations together with normal unfractionated BM cells were resuspended in binding buffer (RPMI-1640 medium buffered at pH 7.4 with 10 mmol/L HEPES and containing 10% [vol/vol] FCS) at a concentration of 1 × 105 to 5 × 108 cells/mL. Resuspended cells were placed in Falcon 2054 tubes (Becton Dickinson & Co, Lincoln Park, NJ) with or without a 100-fold excess of unlabeled cytokine and 10 μL of 125I-cytokine at 105 to 106 cpm/10 μL depending on the dose needed for near saturation of high-affinity receptors. The final volume of each tube was made up to 100 μL with binding buffer. The cells were incubated on ice for 3 hours, after which time cells were resuspended and gently layered on top of a 200 μL cushion of FCS in tapered flexible microcentrifuge tubes (Elkay, Shewsbury, MA). Cell associated (bound) and free 125I-cytokine were separated by centrifugation at 340g for 5 minutes at 4°C. The tip of the tube containing the cell pellet was cut off, and the pellet and supernatant counted in a γ counter (Packard Crystal Multidetector, Packard, Downers Grove, IL) for 1 minute. Saturation isotherms were analyzed using the nonlinear curve fitting program LIGAND22 after correcting for the bindable fraction of the radioactive probe and specific radioactivity.23
The ACK-2 antibody, used to label c-kit+ cells, inhibited binding of 125I-SCF to its receptor, c-kit. To show SCF binding, ACK-2 stained cells were subjected to acid dissociation before performing binding studies. Cells were treated with 3% glacial acetic acid (vol/vol) in ice cold PBS for 1 minute, layered over a FCS cushion, centrifuged at 340g to separate cells from acid, and binding studies performed on the cells as described above.
Cell autoradiography.After 125I-cytokine binding, cell pellets were resuspended in PBS containing 50% (vol/vol) FCS at a final cell concentration of no greater than 5 × 105 cells/mL. Cells (200 μL aliquots) were centrifuged onto gelatin-coated glass slides in a Shandon-Elliot cytocentrifuge, fixed for 5 minutes in ice-cold PBS containing 2.5% (vol/vol) glutaraldehyde, and rinsed three times in distilled water. Slides were air dried overnight, dipped in Kodak NTB2 photographic emulsion (Eastman Kodak, Rochester, NY) at 42°C in a dark room, allowed to dry, sealed in a light-proof box containing drierite and exposed at 4°C for the indicated times, namely SCF, 5 weeks; M-CSF, 4 weeks; IL-3, 8 weeks; GM-CSF, 16 weeks; G-CSF, 6 weeks; IL-6, 24 weeks; LIF, 8 weeks and IL-1α, 16 weeks. Before developing, slides were warmed to room temperature, developed for 3 minutes in Kodak D19 developer (40 g/500 mL of water), washed for 30 seconds in water containing 0.5% (vol/vol) glacial acetic acid, fixed in Agfa G333c fixer (100 mL Part A, 400 mL water containing 0.5% [vol/vol] glacial acetic acid) for 3 minutes, and rinsed in 0.5% (vol/vol) glacial acetic acid in water. Slides were stained in 10% (vol/vol) filtered Giemsa in water for 10 minutes. After drying, slides were mounted in Depex and examined at either 600× or 1,000× magnification. Where possible, grain counts were performed on 100 cells of each morphological type and background grain counts (usually 0 to 3 grains/cell) subtracted.
Conversion of grain counts to receptor numbers.It was assumed that the average number of high-affinity receptors per BM cell, for any given cytokine, would be constant and this was determined for each cytokine by Scatchard analysis of the binding to unfractionated BM cells. In each binding experiment with 125I-cytokine to fractionated cells, unfractionated BM cells were also incubated under identical conditions and exposed to autoradiography under identical conditions to the test samples. After development, the average number of grains per unfractionated BM cell was equated with the average receptor number determined by Scatchard analysis and a value for receptors per grain determined for that experiment. Grains over test cells could then be directly converted to receptors per cell assuming the affinity of receptors in test preparation and or BM cells were the same. For this purpose, low-affinity receptors were not included in the analyses because they are virtually non-detectable by autoradiography (cytokine bound to low-affinity receptors is dissociated during the washing steps, NAN and DM observations).
Reverse transcriptase-PCR (RT-PCR).RNA was extracted from 104 cells using a modified guanidinium thiocyanate method.24 Briefly, cells were lysed and homogenized in 2 mol/L sodium acetate pH 4.0, E coli tRNA (10 μg) and extraction buffer containing 4 mol/L guanidine thiocyanate, 25 mmol/L sodium citrate, 50 mmol/L β-mercaptoethanol, and 0.5% sodium lauroyl sarcosine. After extraction of contaminating proteins in phenol, RNA was precipitated in absolute ethanol and RNA samples were treated with DNase RQ1 (37°C, 30 minutes, Promega, Madison, WI) before reverse transcription. Synthesis of cDNA was performed using random hexamer primers and AMV reverse transcriptase (Boehringer Mannheim, Mannheim, Germany).25
The PCR was performed on a Perkin Elmer Cetus (Norwalk, CT) DNA thermal cycle 480 using the following conditions: 94°C for 1 minute; 56°C for 1 minute; 72°C for 1 minute, for 30 cycles. PCR products were fractionated in 1.2% agarose and transferred to a Zeta Probe membrane (Bio Rad, Richmond, VA).25 Oligonucleotide primers for PCR and probes were as follows: GM-CSFRα, 5′ ATG ACG TCA TCA CAT GCC; 3′ ACG CTG GAT GTC AAT GGC; probe CCC TCA ACC TGA CCT TCG ACC CCT GG: IL-3Rα, 5′ GAG GGA CAT CTT TGT CTG GGC; 3′ GTG ACG TGC ACT ACC GGA T; probe GCT CAT TCC AGT CGC TCT CCC TCT GC: βIL-3 5′ TAC ACA CGA TTT TCT AAT GGA GAT A; 3′ CTA TTC CTA CCC AAC AGC ATC TA; probe CCA GGT CTC CTG GCT TTC ATC AAA GG: βc 5′ ATA CAC GAT TTT CCA TCA CAA ACG; 3′ same as βIL-3; probe GTC AGC ATC AGC TCC TCT GAG GAT CG.26-28 Prehybridization and hybridization was performed as described.29 Positive control cell lines used were the mast cell line MC/9 (obtained from ATCC) and FDC-P1 (obtained from Dr T.M. Dexter, Manchester, UK). The cytotoxic T-lymphocyte cell line (CTLL; obtained from Dr A. Kelso, Brisbane, Australia) is unresponsive to GM-CSF and IL-3 and was used as a negative control for RT-PCR.30
RESULTS
The present studies used three hematopoietic cell populations purified by fluoresence activated cell sorting from mouse BM1; (1) primitive hematopoietic stem cells (Rhlo lin− Ly6A/E+ c-kit+); (2) mature hematopoietic stem cells (Rhhi lin− Ly6A/E+ c-kit+), and (3) progenitor cells (lin− Ly6A/E− c-kit+). Sorted cell populations were incubated with the 125I-cytokine either in the presence or absence of a 100-fold excess of the corresponding unlabeled cytokine, on ice until binding equilibrium was reached (routinely 3 hours). Replicate cytocentrifuge preparations of these cells were then analyzed by autoradiography.
C57BL/6 BM cells were included as a positive control in each series of experiments because they contained subpopulations of cells that express receptors for each of the cytokines studied, and their level of receptor expression was expected to be relatively constant. The data from the analysis using BM cells are shown in Table 1.
Summary of Cytokine Receptors Present on C57BL/6 BM Cells
| 125I-Cytokine . | Av Receptor No./BM Cell* . | % Total BM Labeled . | Reference Cell Population . | % Reference Cells Labeled . | Av Grains/Reference Cell† . | Av Receptor No./Reference Cell . |
|---|---|---|---|---|---|---|
| rr′ SCF | 200 | 17.4 | Blast | 100 | 130 ± 30 | 5,900 |
| rh M-CSF | 76 | 47.3 | PMN | 77 | 27 ± 20 | 80 |
| rm IL-3 | 77 | 61.4 | Monocyte | 100 | 25 ± 19 | 120 |
| rm GM-CSF | 675 | 60.8 | PMN | 100 | 36 ± 9 | 1,100 |
| nm G-CSF | 110 | 51.5 | PMN | 100 | 99 ± 42 | 260 |
| rm IL-6 | 56 | 4.5 | Monocyte | 100 | 59 ± 32 | 990 |
| rh LIF | 6 | 3.6 | Monocyte | 71 | 29 ± 27 | 190 |
| rh IL-1α | nd | nd | PMN | 94 | 14 ± 10 | nd |
| 125I-Cytokine . | Av Receptor No./BM Cell* . | % Total BM Labeled . | Reference Cell Population . | % Reference Cells Labeled . | Av Grains/Reference Cell† . | Av Receptor No./Reference Cell . |
|---|---|---|---|---|---|---|
| rr′ SCF | 200 | 17.4 | Blast | 100 | 130 ± 30 | 5,900 |
| rh M-CSF | 76 | 47.3 | PMN | 77 | 27 ± 20 | 80 |
| rm IL-3 | 77 | 61.4 | Monocyte | 100 | 25 ± 19 | 120 |
| rm GM-CSF | 675 | 60.8 | PMN | 100 | 36 ± 9 | 1,100 |
| nm G-CSF | 110 | 51.5 | PMN | 100 | 99 ± 42 | 260 |
| rm IL-6 | 56 | 4.5 | Monocyte | 100 | 59 ± 32 | 990 |
| rh LIF | 6 | 3.6 | Monocyte | 71 | 29 ± 27 | 190 |
| rh IL-1α | nd | nd | PMN | 94 | 14 ± 10 | nd |
All labeling was completely inhibited in the presence of a 100-fold excess of the appropriate unlabeled cytokine.
Abbreviation: nd, not determined.
Determined by Scatchard analysis on 5 × 106 C57BL/6 BM cells incubated on ice for 3 hours.
Mean grain counts ± standard deviations of labeled cells.
The percentage of cells labeled within each sorted cell population and the average number of silver grains per labeled cell for each radioiodinated cytokine investigated are summarized in Table 2. In calculating receptor numbers from grain counts it was assumed that the average grain count per BM cell is proportional to the average number of high-affinity receptors determined by Scatchard analysis of the same population. However, this is only true if the affinity of the receptor on each cell type is similar and if receptors are detected equally by both techniques. This proportionality does not apply for low-affinity receptors with fast dissociation-rates because autoradiography involves several washing steps (where radioactive ligand may dissociate) whereas Scatchard analysis in our hands, involves only one rapid separation step. The cytokine receptor numbers calculated to be present on stem and progenitor cell populations are presented in Table 2.
Labeling of Highly Enriched Populations of Primitive and Mature Hemopoietic Stem Cells and Progenitor Cells
| 125I-Cytokine . | Primitive HSC* Rhlolin−Sca-1+c-kit+ . | Mature HSC* Rhhilin−Sca-1+c-kit+ . | Progenitors* lin−Sca-1−c-kit+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . |
| rr′ SCF | † | † | 17† | 21 ± 29 | 960 | ||||
| rh M-CSF | 3 | 30 ± 29 | 90 | 0 | 0 | 0 | 0 | 0 | 0 |
| rm IL-3 | 65 | 16 ± 12 | 80 | 69 | 24 ± 15 | 110 | 93 | 24 ± 24 | 110 |
| rm GM-CSF | 0 | 0 | 0 | 3 | 6 ± 4 | 190 | 55 | 18 ± 14 | 560 |
| nm G-CSF | 79 | 22 ± 21 | 60 | 73 | 30 ± 22 | 80 | 93 | 10 ± 11 | 30 |
| rm IL-6 | 13 | 10 ± 2 | 170 | 18 | 10 ± 5 | 170 | 22 | 24 ± 14 | 400 |
| rh LIF | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 ± 1 | 30 |
| rh IL-1α | 56 | 5 ± 2 | nd | 64 | 17 ± 11 | nd | 43 | 6 ± 3 | nd |
| 125I-Cytokine . | Primitive HSC* Rhlolin−Sca-1+c-kit+ . | Mature HSC* Rhhilin−Sca-1+c-kit+ . | Progenitors* lin−Sca-1−c-kit+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . | % Cells Labeled . | Av Grains/Labeled Cell‡ . | Receptors/Labeled Cell . |
| rr′ SCF | † | † | 17† | 21 ± 29 | 960 | ||||
| rh M-CSF | 3 | 30 ± 29 | 90 | 0 | 0 | 0 | 0 | 0 | 0 |
| rm IL-3 | 65 | 16 ± 12 | 80 | 69 | 24 ± 15 | 110 | 93 | 24 ± 24 | 110 |
| rm GM-CSF | 0 | 0 | 0 | 3 | 6 ± 4 | 190 | 55 | 18 ± 14 | 560 |
| nm G-CSF | 79 | 22 ± 21 | 60 | 73 | 30 ± 22 | 80 | 93 | 10 ± 11 | 30 |
| rm IL-6 | 13 | 10 ± 2 | 170 | 18 | 10 ± 5 | 170 | 22 | 24 ± 14 | 400 |
| rh LIF | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 ± 1 | 30 |
| rh IL-1α | 56 | 5 ± 2 | nd | 64 | 17 ± 11 | nd | 43 | 6 ± 3 | nd |
All labeling was completely inhibited in the presence of a 100-fold excess of the appropriate unlabeled cytokine.
Abbreviation: nd, not determined because Scatchard analysis was not performed for rh IL-1α.
Sca-1 is identical to Ly6A/E.
Anti c-kit antibody (ACK2) inhibited binding of SCF to c-kit–positive cells. This effect could be partially reversed by acid treating the cells before performing SCF binding.
Mean grain counts ± standard deviations of labeled cells.
SCF.Within marrow populations, prominent labeling was observed of approximately 100% blast cells (5,900 receptors per cell), which accounted for two-thirds of the total binding to marrow cells, and this labeling was completely blocked by coincubation with a 100-fold excess of unlabeled SCF. Labeling of other cell types within the BM was similar to that reported previously from an analysis of BALB/c marrow cells instead of C57BL/6 to characterize 125I-rr′-SCF.31 In addition to heavily labeled blast cells, the majority of promyelocytes and myelocytes were labeled but grain counts were very low. Few metamyelocytes and no polymorphonuclear neutrophils (PMNs) were labeled. Seven percent of lymphocyte-like cells were heavily labeled and no labeling was observed of nucleated erythroid cells. Both HSC populations and the progenitor cell population were sorted on the basis of positive expression for c-kit so it would be expected that these populations would bind 125I-rr′-SCF. However, initial experiments revealed that these populations were not labeled or had very few grains per cell presumably because the antibody against c-kit (ACK-2) used in the fractionation procedure blocked subsequent binding of 125I-rr′-SCF to antibody-treated cells. This blocking effect on progenitor cells was partially reversed by acid treatment of the cells to remove antibody before binding. Unfortunately, following acid treatment of the HSC populations, no cells were recovered on both occasions tested because of the small number of cells used and the relative ease with which acid-treated cells tended to stick to plastic surfaces.
M-CSF.Within the BM population, unambiguous labeling was observed of 100% of monocytes (>200 grains/cell) and 77% of PMNs (mean grain count of 27 grains per cell being equivalent to 80 receptors per labeled PMN). This pattern of labeling is in partial agreement with the findings reported previously32 with the exception that labeled polymorphs were observed. Cold competion with 100-fold excess of unlabeled M-CSF completely blocked the labeling of both monocytes and PMNs. No labeling was observed of either stem cell population, suggesting that stem cells do not exhibit receptors for M-CSF. However, more puzzling was the finding that no receptors were detected on the highly enriched progenitor cell population which is known to respond to M-CSF in in vitro colony-forming assays.
IL-3.Autoradiographic analysis of BM revealed 100% of monocytes were heterogeneously labeled (average grain count of 25 ± 19 being equivalent to 120 high-affinity receptors per cell). In addition, prominent labeling was observed of 100% of promonocytes (350 receptors/labeled cell), 88% of blast cells (330 receptors/labeled cell), 100% of promyelocytes (450 receptors/labeled cell), 100% of myelocytes (300 receptors/labeled cell), 100% of metamyelocytes and PMNs (170 receptors/labeled cell), 100% of eosinophils (340 receptors/labeled cell), and a small subpopulation of lymphocyte-like cells (7%). In all cases labeling was completely blocked by co-incubation with a 100-fold excess of unlabeled rm-IL-3. No labeling was detected on nucleated erythroid cells. The observed labeling of BM agreed with the data from previous studies using BALB/c BM cells.33 125I-rm-IL-3 labeling was detected on both stem cell populations; 65% of Rhlolin− Ly6A/E+ c-kit+ (80 receptors/labeled cell) and 69% of Rhhi lin− Ly6A/E+ c-kit+ (110 receptors/labeled cell). Ninety-three percent of the purified progenitor cells were labeled (110 receptors/labeled cell).
GM-CSF.Within the C57BL/6 BM populations, prominent labeling was observed of 100% metamyelocytes and PMNs (1,100 receptors/labeled cell), 100% of promyelocytes (2,000 receptors/labeled cell), 100% of monocytes (860 receptors/labeled cell), 90% of blast cells (1,400 receptors/labeled cell), and 93% of eosinophils (520 receptors/labeled cell). Labeling was completely blocked with a 100-fold excess of unlabeled rm-GM-CSF. No significant labeling was observed on either stem cell population, whereas 55% of the progenitor cell population was labeled with GM-CSF (560 receptors/labeled cell).
G-CSF.As described previously34 labeling of C57BL/6 BM cells with 125I-nm-G-CSF revealed that 100% of PMNs were labeled with 99 ± 42 silver grains per cell (equivalent to an average of 260 receptors/labeled cell), 76% of blast cells (160 receptors/cell), and 94% of promyelocyte/myelocytes were labeled (200 receptors/cell). Very few receptors were detected on monocytes at any stage of differentiation. G-CSF binding was detected on both stem cell populations; 79% of Rhlolin− Ly6A/E+ c-kit+ (60 receptors/labeled cell) and 73% of Rhhilin− Ly6A/E+ c-kit+ (80 receptors/labeled cell) were labeled. Labeling of 93% of the progenitor cell population (30 receptors/labeled cell) was also observed. Labeling of all cell types was completely blocked with the incubation of 100-fold excess of unlabeled nm-G-CSF.
IL-6.In agreement with previous studies,35 C57BL/6 BM cells labeled with 125I-rm-IL-6 showed unambiguous labeling of monocytes (100%) with a mean grain count of 59 ± 32 per labeled cell (990 receptors/cell). Labeling was also observed on approximately 10% of undifferentiated blast cells (1,700 receptors/labeled cell). No labeling was observed of PMNs, eosinophils, nucleated erythroid cells, or the majority of marrow lymphocytes. With purified stem cells, 13% of the Rhlolin− Ly6A/E+ c-kit+ population (170 receptors/labeled cell) and 18% Rhhilin− Ly6A/E+ c-kit+ population (170 receptors/labeled cell) were labeled. In addition, 22% of the progenitor cell population was labeled (400 receptors/labeled cell). The labeling of marrow and sorted cell populations was completely blocked by co-incubation with a 100-fold excess of unlabeled rm-IL-6.
LIF.The distribution of LIF receptors on murine BM and the sorted cell populations used 125I-rh-LIF rather than 125I-rm-LIF, because studies36 reported human LIF bound the murine LIF receptor with the same specificity and affinity as murine LIF but had a higher specific radioactivity. Within C57BL/6 BM cells, labeling was restricted to cells of the monocyte/macrophage lineage (71% labeled; 190 receptors/labeled cell) and a small subset (8%) of lymphocyte-like cells (90 receptors/labeled cell). Labeling was not detected on neutrophils and eosinophils at any stage of differentiation, nucleated erythroid cells, mast cells or the majority of lymphocytes. Previous studies reported a similar distribution of LIF receptors on BM cells.37 No receptors were observed for LIF on either of the HSC populations, but 1% of the progenitor cell population (30 receptors/labeled cell) was labeled. The labeling of BM and sorted cell populations was completely blocked by coincubation with a 100-fold excess of unlabeled LIF.
IL-1α.Autoradiographic analysis revealed that 94% of C57BL/6 BM PMNs were labeled with an average grain count per labeled cell of 14 ± 10. Labeling was also observed on 48% of blast cells (7 ± 4 silver grains per labeled cell), 80% of promyelocytes/myelocytes (9 ± 4 grains/cell), 50% of promonocytes (19 ± 22 grains/cell), and 54% of monocytes (11 ± 11 grains/cell). With purified stem cells 56% (5 ± 2 grains/cell) and 64% (17 ± 11 grains/cell) of the primitive and mature HSC populations were labeled, respectively, as was 43% of the progenitor cell population (6 ± 3 grains/cell). Labeling of BM and sorted cell populations was completely blocked by co-incubation with a 100-fold excess of unlabeled IL-1α.
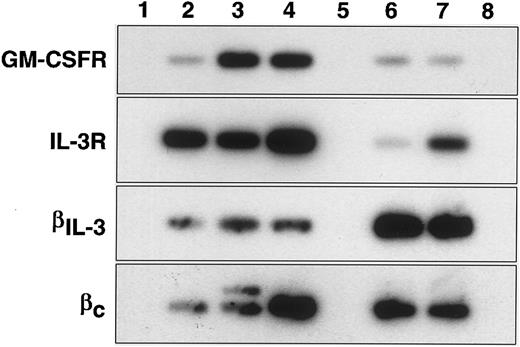
PCR analysis of GM-CSF and IL-3 receptor message expression.In confirmation of the direct binding studies, the IL-3 receptor α-chain mRNA was expressed in all three primitive cell populations (stem cell and progenitor cell populations). In contrast GM-CSF receptor α-chain mRNA was only very weakly expressed in the primitive HSC population but there was increased expression in the mature HSC and progenitor cell populations. This is only partially in agreement with the direct binding studies and may suggest that functional high affinity receptors for GM-CSF do not form in the mature HSC population despite the presence of mRNA for the α-chain. Since high affinity IL-3 receptors can use either βc or βIL-3 chains along with the α-chain, it was not clear from the binding studies which β-chains were expressed in the stem cell compartment. Figure 1 shows that βIL-3 and βc were both expressed in the two stem cell populations and that βc-, but not βIL-3-chain, expression may have been elevated in the progenitor cell population.
RT-PCR analysis of CSF receptors in primitive cells. Agarose-gel fractionated RT-PCR products of GM-CSF receptor α (GM-CSFR), interleukin-3 receptor α (IL-3R), β-chain of IL-3 receptor (βIL-3) and common β-chain of IL-3, IL-5 and GM-CSF receptors (βc) were transferred to nylon membranes and hybridized with internal radioactive oligonucleotides. Lane numbers refer to: (1) “no reverse transcriptase” control, (2) Rhlo lin− Ly6A/E+ c-kit+, (3) Rhhi lin− Ly6A/E+ c-kit+, (4) lin− Ly6A/E− c-kit+, (5) blank, (6) MC/9 cells, (7) FDC-P1 cells, (8) CTLL cells.
RT-PCR analysis of CSF receptors in primitive cells. Agarose-gel fractionated RT-PCR products of GM-CSF receptor α (GM-CSFR), interleukin-3 receptor α (IL-3R), β-chain of IL-3 receptor (βIL-3) and common β-chain of IL-3, IL-5 and GM-CSF receptors (βc) were transferred to nylon membranes and hybridized with internal radioactive oligonucleotides. Lane numbers refer to: (1) “no reverse transcriptase” control, (2) Rhlo lin− Ly6A/E+ c-kit+, (3) Rhhi lin− Ly6A/E+ c-kit+, (4) lin− Ly6A/E− c-kit+, (5) blank, (6) MC/9 cells, (7) FDC-P1 cells, (8) CTLL cells.
DISCUSSION
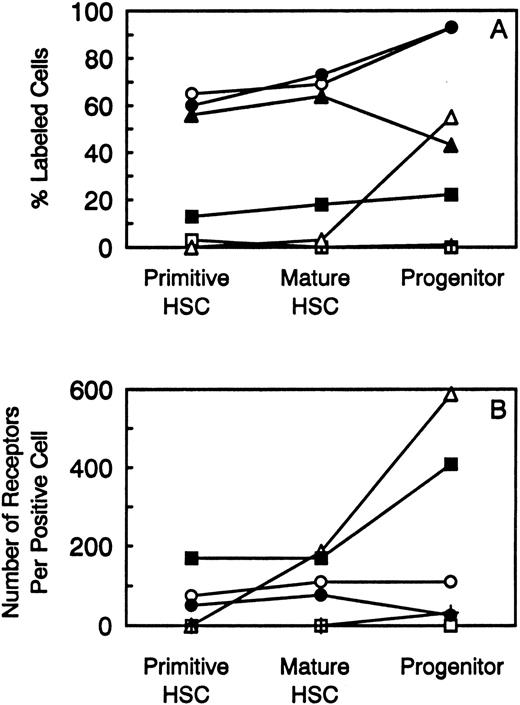
The results presented in this report describe the cytokine receptor profile on highly enriched populations of hematopoietic stem and progenitor cells as summarized in Fig 2. Analyses of cytokine receptors on both primitive and mature HSC populations revealed similar profiles. Both populations can be assumed to express receptors for SCF, because they were sorted according to their expression of c-kit. Although only few SCF receptors were detected by autoradiography, it was shown that the antibody against c-kit, ACK-2, inhibited SCF binding. This effect could be reversed by acid dissociation of the antibody before performing binding studies. The expression of c-kit has previously been used by other investigators to sort for both murine and human HSCs.5,6 38-42
Summary of the percentage of stem and progenitor cell populations labeled with 125I-cytokine (A) and the number of receptors expressed on positive cells (B). The following symbols represent the cytokine receptors analyzed: (□) M-CSF; (▪) IL-6; (○) IL-3; (•) G-CSF; (▵) GM-CSF; (▴) IL-1; and (+) LIF.
Summary of the percentage of stem and progenitor cell populations labeled with 125I-cytokine (A) and the number of receptors expressed on positive cells (B). The following symbols represent the cytokine receptors analyzed: (□) M-CSF; (▪) IL-6; (○) IL-3; (•) G-CSF; (▵) GM-CSF; (▴) IL-1; and (+) LIF.
Cells within both stem cell populations expressed receptors for IL-3, G-CSF, IL-6, and IL-1. No receptors were detected for M-CSF, GM-CSF, or LIF although the BM controls showed successful labeling of cell populations. Previous studies1 revealed that these HSCs proliferate and differentiate in vitro in response to multiple cytokine combinations (SCF, IL-1, IL-3, IL-6, G-CSF, GM-CSF, and M-CSF ) but very little mitogenic/survival response was observed in the presence of single cytokines. The inability to detect receptors for GM-CSF and M-CSF on HSCs may be in part explained by the ability of combinations of SCF, IL-1, IL-3, IL-6, and G-CSF to stimulate the differentiation of HSCs to progenitor cells which is correlated with expression of GM-CSF and M-CSF receptors. RT-PCR analysis by others of cytokine receptor expression in a “purified” HSC population (Rhlo wheat-germ agglutinin+) depleted of monocytes and macrophages revealed expression of mRNA for c-kit. However, very few cell surface receptors were detected as assessed by their ability to bind biotinylated SCF.43 In partial contrast to our results, mRNA transcripts were detected for βIL-3 but not βc or flk-2. However, in their study, data were not provided regarding the purity of this population or the ability of these cells to contribute to long-term BM repopulation. Studies using early human progenitor cells purified from peripheral blood and enriched for burst-forming unit erythroid, colony-forming unit granulocyte macrophage (CFU-GM), and CFU-GEMM revealed that high-affinity receptors were present for IL-3, IL-6, GM-CSF, and EPO.44 However, the “stem cell” populations used in their studies were highly heterogeneous and, since only Scatchard analysis was performed, no conclusions could be drawn as to the percentage of the population that contributed to the binding observed.
The receptors for SCF and IL-3 demonstrated on purified stem cells and progenitor cells may explain in part why a small subset of lymphocyte-like cells in unfractionated marrow populations is always observed to express receptors for these cytokines since no cells in lymphoid organs express such receptors.
Although receptors for GM-CSF were detected on the progenitor cell population in addition to receptors for SCF, IL-1, IL-3, IL-6, and G-CSF, no receptors were detected for M-CSF. The presence of receptors for GM-CSF on progenitor cells clearly represents increased expression of receptor relative to stem cells both in terms of the percentage of the population labeled and in the number of receptors per positive cell. The absence of receptors for M-CSF (c-fms) on the highly enriched progenitor cell population is somewhat confusing as these cells were capable of responding to M-CSF in in vitro colony forming cell assays. The loss of c-fms from these cells may be explained by the ability of the common contaminant, bacterial endotoxin (LPS), to rapidly downregulate c-fms expression.45-47 Alternatively, c-fms expression may be induced on the lin− Ly6A/E− c-kit+ population through the effects of culture in vitro.
Within both the HSC and progenitor cell populations, heterogeneity was detected in terms of the percentage of the population expressing receptors and the number of receptors expressed by labeled cells. This heterogeneity suggests that cytokine receptor expression could be used to further subdivide HSC populations either by using the highly sensitive technique of flow cytometry utilizing three-layer immunofluorescence that is capable of detecting cells expressing as few as 100 receptors per cell,48 or by methods employing toxins (that catalytically inactivate protein synthesis) linked either chemically or genetically to cytokines and/or cytokine receptor antibodies could be used to deplete receptor-positive cell populations.49 Pure populations of HSCs would be useful in BM transplantation and as vehicles for correcting genetic abnormalities such as the thalassaemias and severe combined immunodeficiency syndromes.
ACKNOWLEDGMENT
The authors acknowledge the expert technical assistance of Shan-Li Liu for preparing BM cell suspensions, and Dr Francis Battye and Ralph Rossi for help with FACS.
Submitted May 28, 1996; accepted August 9, 1996.
Supported by the Anti-Cancer Council of Victoria, The National Health and Medical Research Council of Australia; the J.D. and L. Harris Trust Fund; the Philip Bushell Trust; the National Institutes of Health Grant No. CA22556; and the Australian Government Co-operative Research Centres Scheme.
Address reprint requests to Donald Metcalf, MD, The Walter and Eliza Hall Institute of Medical Research, Post Office Royal Melbourne Hospital, 3050 Victoria, Australia.
REFERENCES
Author notes
Deceased.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal