Abstract
CD34+ hematopoietic progenitor cells were assessed for mRNA expression of the human immunodeficiency virus type-1 (HIV-1) coreceptors CXCR-4, also termed fusin or LESTR, and CKR-5, also called CC-CKR-5 or CCR-5. The CD34+ cells were obtained from leukapheresis products of 17 patients after granulocyte colony-stimulating factor–supported cytotoxic chemotherapy. Using a two-step enrichment procedure including immunomagnetic bead separation and fluorescence-activated cell sorting, the CD34+ cells had a median purity of 99.8%. Assessing 9 CD34+ cell samples by polymerase chain reaction after reverse transcription (RT-PCR), CXCR-4 mRNA was found in all samples, whereas CKR-5 mRNA was only present in 3 samples, even though a nested PCR was used. Eight additional CD34+ cell samples were sorted according to CD4 expression. Based on a three-color immunofluorescence analysis, the mean relative fluorescence intensity of HLA-DR was smaller on CD34+/CD4+ cells in comparison with CD34+/CD4− cells. CXCR-4 mRNA was found in 5 of 8 CD34+/CD4+ samples and in 7 of 8 CD34+/CD4− samples, whereas CKR-5 mRNA was detected in 2 CD34+/CD4+ samples and in 1 CD34+/CD4− cell sample. Looking at the total number of CD34+ cell samples examined, the proportion of specimens containing CXCR-4 mRNA was 84% in comparison with 24% of specimens positive for CKR-5 mRNA. These data suggest that CD34+/CD4+ hematopoietic progenitor cells, including true stem cell candidates, could be susceptible to HIV-1 infection. Considering the relatively low incidence of CD34+ cell samples containing CKR-5 mRNA, CD34+/CD4+ cells appear to be particularly prone for HIV-1 infection via the CXCR-4 coreceptor. Because this chemokine receptor allows T-cell–tropic HIV-1 strains to infect cells, CD34+ cells expressing CD4 and CXCR-4 might be infected by HIV-1 during later stages of the disease, following a viral phenotype switch from macrophage- to T-cell–tropic HIV-1 strains.
CYTOPENIAS IN THE peripheral blood, associated with myelodysplastic changes in the bone marrow, are observed in 40% to 80% of patients with the acquired immunodeficiency syndrome (AIDS).1 There is some evidence that CD34+ hematopoietic progenitor and stem cells are affected by the human immunodeficiency virus type-1 (HIV-1). In short-term cultures, the clonal growth of hematopoietic progenitor cells from bone marrow or peripheral blood of patients with HIV-1 infection was impaired.2-6 Using polymerase chain reaction (PCR), HIV-1 proviral DNA was only rarely found in purified CD34+ bone marrow cells from asymptomatic patients with HIV-1 infection,7,8 whereas CD34+ bone marrow cells of patients with AIDS contained the HIV-1 genome to some extent.5 9
In vitro, bone marrow-derived CD34+ cells were susceptible to HIV-1 infection.10-12 Between 9% and 23% of purified CD34+ cells could be infected by T-cell–tropic HIV-1 as shown in single-cell cultures.13 Hematopoietic progenitor cells are potential target cells for HIV-1, because 25% to 50% of the CD34+ cells coexpress CD4, which serves as high-affinity receptor for HIV-1.14,15 This cell surface antigen is predominantly expressed on phenotypical more primitive progenitor cells and might regulate the interaction with major histocompatibility class-II–positive bone marrow accessory cells such as macrophages, B cells, or activated T cells.14
Recently, two chemokine receptors were identified that are required as coreceptors for HIV-1 infection. CXCR-4, also termed as fusin or LESTR, functions as a coreceptor for T-cell–tropic HIV-1 strains.16 Because of sequence homology with and chromosomal mapping near other CXC chemokine receptor genes,17-19 fusin recently was renamed CXCR-4.20 On the other hand, CKR-5, also called CC-CKR5 or CCR-5, is used by macrophage-tropic HIV-1 strains.21-24
In this report, we examined highly enriched CD34+ and CD34+/CD4+ hematopoietic progenitor cells from 17 individuals for mRNA expression of HIV-1 coreceptors CXCR-4 and CKR-5. To obtain a sufficient number of purified CD34+ cells, blood-derived mononuclear cells were collected by leukapheresis from cancer patients after granulocyte colony-stimulating factor (G-CSF )–supported cytotoxic chemotherapy. Because monoclonal antibodies (MoAbs) directed against CXCR-4 or CKR-5 for immunphenotyping are not available yet, we assessed mRNA expression using PCR following reverse transcription (RT-PCR).
MATERIALS AND METHODS
Cell lines and plasmids.The human cell lines CEM (acute lymphocytic leukemia), HL60 (promyelocytic leukemia), K562 (erythroleukemia), U-87MG (glioblastoma/astrocytoma), and SK-N-MC (neuroblastoma) were obtained from the tumor cell collection of the German Cancer Research Center (Heidelberg, Germany). Human cell lines KG1a (promyeloblastic leukemia) and HUT-78 (T-cell lymphoma) were obtained from American Type and Tissue Culture Collection (ATTC; Rockville, MD).
Using Northern blot hybridization, HL60 was found to be positive for CXCR-4 mRNA,16,18 whereas U-87MG and SK-N-MC are negative.17,19 CKR-5 mRNA is present in KG1a, but is not expressed in HL60 or K562.25 For the cell line HUT-78, the data on CKR-5 transcription are controversial.21 22
Plasmid pcDNA3-LESTR, containing CXCR-4 cDNA cloned in pcDNA3 vector (Invitrogen, Carlsbad, CA), was kindly provided by M. Loetscher (Theodor Kocher Institut, Bern, Switzerland).19 Plasmid pcDNA3/ChemR13, containing the coding region of CC-CKR5, was kindly provided by M. Parmentier (ULB Campus Erasme, Bruxelles).25
Collection of blood mononuclear cells.Blood mononuclear cells were obtained after informed consent from 17 adult patients with solid tumors or hematologic malignancies (4 breast cancer, 7 lymphoma, 4 multiple myeloma, 1 ovarian carcinoma, and 1 chorioncarcinoma). All patients received cytotoxic chemotherapy followed by recombinant human G-CSF (R-metHuG-CSF [filgrastim]; Amgen, Thousand Oaks, CA; 300 μg/d subcutanously). At the time of leukapheresis, all patients were in remission. The median age was 42 years (range, 26 to 56 years). During bone marrow recovery, peripheral blood mononuclear cells were collected by leukapheresis using a Fenwal CS 3,000 (Baxter Deutschland, München, Germany).
A median number of 6.6 × 108 mononuclear cells (range, 2.8 × 108 to 16 × 108 cells) from leukapheresis products were diluted in 30 mL RPMI 1640 medium (CC Pro, Neustadt/Weinstrasse, Germany), underlayed with 15 mL Lymphoprep (Nycomed Pharma, Oslo, Norway) and centrifuged at 870g for 20 minutes. The cells of the interface were removed and resuspended in 30 mL of RPMI 1640. The cells were centrifuged at 630g for 10 minutes, followed by a second washing procedure.
CD34+ cell purification by immunomagnetic beads.CD34+ cells from leukapheresis products were positively selected using the MiniMACSR immunomagnetic separation system (Milteny Biotec, Bergisch Gladbach, Germany).26 According to the manufacturer's instructions, 108 mononuclear cells were suspended in 300 μL sorting buffer (1× phosphate-buffered saline supplemented with 2 mmol/L EDTA and 0.5% bovine serum albumine), 100 μL human Ig FcR blocking antibody, and 100 mL monoclonal hapten-conjugated CD34 antibody (clone QBEND/10; Milteny Biotec) and incubated for 15 minutes at 4°C. Cells were washed with 15 mL sorting buffer and centrifuged at 700g for 10 minutes. The pellet was resuspended in 400 μL sorting buffer, and cells were incubated with 100 μL paramagnetic microbeads conjugated to antihapten antibody (Milteny Biotec) per 108 cells for 15 minutes at 4°C. Cells were washed, resuspended in sorting buffer, passed through a 30-μm nylon mesh, and separated in a column exposed to the magnetic field of the MACS device. The column was washed four times with 500 μL sorting buffer and removed from the separator, and the retained cells were eluted with 1 mL sorting buffer using a plunger. Eluted cells were subjected to a second round of separation.
Fluorescence activated cell sorting (FACS).The selected CD34+ cells were stained for 30 minutes at 4°C with fluorescein isothiocyanate (FITC)-conjugated MoAb HPCA (CD34), phycoerythrin (PE)-conjugated antibody Leu-3a (CD4; Becton Dickinson, San Jose, CA), and PE/Cy5-conjugated antibody HLA-DR (Clone Immu-357; Coulter-Immunotech, Hamburg, Germany). Isotype-identical MoAbs served as control (IgG1-FITC and IgG1-PE [Becton Dickinson]; IgG2a-PE/Cy5 [Coulter-Immunotech]). Cells were analyzed and sorted on a FACSVantage cell sorter using a 150-mW single argon laser at 488 nm (Becton Dickinson). Fluorescence was measured using 530/15 nm (FITC), 575/26 nm (PE), and 675/36 nm (PE/Cy5) band pass filters. Gates for cell sorting were set on CD34+, CD34+/CD4+, and CD34+/CD4− cells, respectively. Purity of sorted cells was assessed by flow cytometry.
RNA extraction and reverse transcription.Total cellular RNA was isolated as described.27 Briefly, cells were washed twice in ice-cold phosphate-buffered saline and resuspended in 500 μL ice-cold RNA extraction buffer (4 mol/L guanidinium thiocyanate, 25 mmol/L sodium citrate, pH 7, 0.5% sarcosyl, and 0.1 mol/L 2-mercaptoethanol). Fifty microlilters of 2 mol/L sodium acetate, pH 4, 500 μL water-saturated phenol, and 100 μL chloroform-isoamyl alcohol (49:1) were added. The suspension was shaken vigorously, cooled on ice for 15 minutes, and centrifuged at 10,000g for 20 minutes. RNA in aqueous phase was precipitated with 1 mL isopropanol at −20°C for 1 hour and sedimentated at 10,000g for 20 minutes. RNA pellet was resuspended in 300 μL RNA extraction buffer without mercaptoethanol. After the addition of 300 μL isopropanol, RNA was incubated at −20°C for 1 hour and centrifuged at 10,000g for 20 minutes. The RNA pellet was resuspended in 75% ethanol, centrifuged, vacuum dried, and dissolved in 10 mmol/L Tris-HCl, pH 8, 1 mmol/L EDTA.
RNA concentrations were measured using a GeneQuant II spectrophotometer (Pharmacia, Cambridge, UK). For elimination of contaminating genomic DNA, RNA was digested using 4 U RQ1 RNase free DNase (Promega, Madison, WI) at 37°C for 90 minutes in a final volume of 28 μL containing 70 mmol/L Tris-HCl, pH 8.3, 107 mmol/L KCl, 4 mmol/L MgCl2 , and 40 U RNAsin (Promega). Fourteen microliters of samples, containing 50 to 1,000 ng RNA, were heated at 65°C for 5 minutes and reversely transcribed for 30 minutes at 37°C using 200 U reverse transcriptase (GIBCO BRL, Paisley, UK). The final volume of 22 μL contained 0.5 mmol/L dATP, dTTP, dGTP, and dCTP each; 1 μmol/L random hexamers (Boehringer Mannheim, Mannheim, Germany); 10 mmol/L dithiothreitol; 40 U of RNAsin; 45 mmol/L Tris-HCl, pH 8.3; 70 mmol/L KCl; and 3 mmol/L MgCl2 . Samples were denatured at 94°C for 2 minutes and stored at −20°C. Chemicals were obtained from Merck (Darmstadt, Germany) and Sigma (Deisenhofen, Germany).
PCR.PCR was performed in 50 μL containing 20 mmol/L Tris-HCl, pH 8.4; 50 mmol/L KCl; 0.2 mmol/L dATP, dTTP, dGTP, and dCTP each; 1.5 mmol/L MgCl2 ; 2.5 U Taq DNA polymerase (GIBCO BRL); 5 μL cDNA; and 1 pmol of each primer. A denaturation step lasting 2 minutes and 45 seconds was followed by 40 cycles of amplification with a PCR processor (GeneAmp 2400; Perkin Elmer, Norwalk, CT). Each cycle included a denaturation step of 20 seconds at 95°C, a primer annealing step of 30 seconds, and a chain elongation step of 30 seconds at 72°C. The final elongation step was prolonged to 7 minutes to ensure a complete extension of the amplified DNA. The sequences of the primers were as follows: GAPDH-SE-487, 5′-TCC ATG ACA ACT TTG GTA TCG-3′; GAPDH-AS-863, 5′-GTC GCT GTT GAA GTC AGA GGA-3′; CXCR-4-SE-527, 5′-ACG TCA GTG AGG CAG ATG-3′; CXCR-4-SE-553, 5′-ATC TGT GAC CGC TTC TAC-3′; CXCR-4-AS-729, 5′-GAT GAC TGT GGT CTT GAG-3′; CKR-5-SE-259, 5′-GTC CAA TCT ATG ACA TCA-3′; CKR-5-SE-279, 5′-TAT TAT ACA TCG GAG CCC-3′; CKR-5-AS-770, 5′-GGT GTA ATG AAG ACC TTC-3′; and CKR-5-AS-806, 5′-GAA TTG ATA CTG ACT GTA-3′. CXCR-4 cDNA was amplified using primer pair CXCR-4-SE-527/CXCR-4-AS-729, whereas CKR-5 cDNA was amplified in a nested PCR using the primer pairs CKR-5-SE-259/CKR-5-AS-806 and CKR-5-SE-279/CKR-5-AS-770. Annealing temperatures were 60°C (GAPDH-SE-487/GAPDH-AS-863), 64°C (CXCR-4-SE-527/CXCR-4-AS-729), and 55°C (CKR-SE-259/CKR-AS-806, CKR-SE-279/CKR-AS-770). From each sample, PCRs amplifying glycerylaldehyde-6-phosphate dehydrogenase (GAPDH) cDNA were performed to confirm the presence of amplifyable cDNA after reverse transcription.28 Additionally, from each RNA sample, PCR was performed without reverse transcription to exclude genomic DNA contamination. PCR products were analyzed by 1.8% agarose gel electrophoresis.
Hybridization of PCR products.For Southern hybridization, amplified CXCR-4 and CC-CKR5 PCR products were separated on agarose gels and transferred to nylon membranes (GeneScreen Plus; Du Pont, Boston, MA) as described.29 Radioactive labeled pcDNA3-LESTR and pcDNA3/ChemR13 DNA were used as probes for hybridization of CXCR-4 and CKR-5, respectively. Fifty nanograms of denatured DNA was labeled with 25 μCi [α-32P]dCTP using 3 U Klenow Enzyme (Boehringer Mannheim); 20 μmol/L dATP, dTTP, and dGTP each; 5 U random hexadeoxynucleotides; 20 μg bovine serum albumine; 0.2 mol/L HEPES; and 15 mmol/L Tris-HCl, pH 8, in a final volume of 20 μL at 37°C for 30 minutes. Hybridization was performed overnight at 42°C in a solution containing 50% formamide, 0.75 mol/L NaCl, 0.075 mol/L sodium citrate, pH 7.0, 100 μg t-RNA/mL, 0.02 mol/L sodium pyrophosphate, 1% sodium lauryl sulfate, and 1× Denhardt's solution (0.02% each bovine serum albumine, Ficoll, and polyvinylpyrrolidone). After hybridization, filters were washed three times, for 30 minutes each time, with 0.3 mol/L sodium chloride, 0.03 mol/L sodium citrate, and 0.1% sodium lauryl sulfate at 68°C and exposed to x-ray film (NEF 496; DuPont).
Sequencing of PCR products.PCR products amplified with primers LESTR-SE-527/LESTR-AS-729 and CKR-SE-259/CKR-AS-806 were purified using QIAquick columns (Qiagen, Chatsworth, CA). DNA fragments were sequenced using the internal primers LESTR-SE-573 and CKR-SE-279, respectively, and [α-35S]dATP in CircumVent thermal cycle sequencing reactions (Biolabs, Beverly, CA).30 Sequencing was performed in 20 cycles including a denaturation step at 95°C for 20 seconds, a primer annealing step at 55°C for 20 seconds, and a chain elongation step at 72°C for 20 seconds. The samples were analyzed by polyacrylamide gel electrophoresis under denaturing conditions (9% polyacrylamide gels containing 7 mol/L urea in 89 mmol/L Tris-borate buffer, pH 8.3). Gels were dried and exposed to x-ray film (Hyperfilm β-max; Amersham, Arlington Heights, IL). Sequences of PCR products were compared with CXCR-4 cDNA (EMBL data bank accession no. X71635) and CKR5 cDNA sequences (EMBL data bank accession no. X91492) using the softwares Clustal and BESTfit of the Heidelberg Unix Sequence Analysis Resources (HUSAR, version 4.0; DKFZ, Heidelberg, Germany).
RESULTS
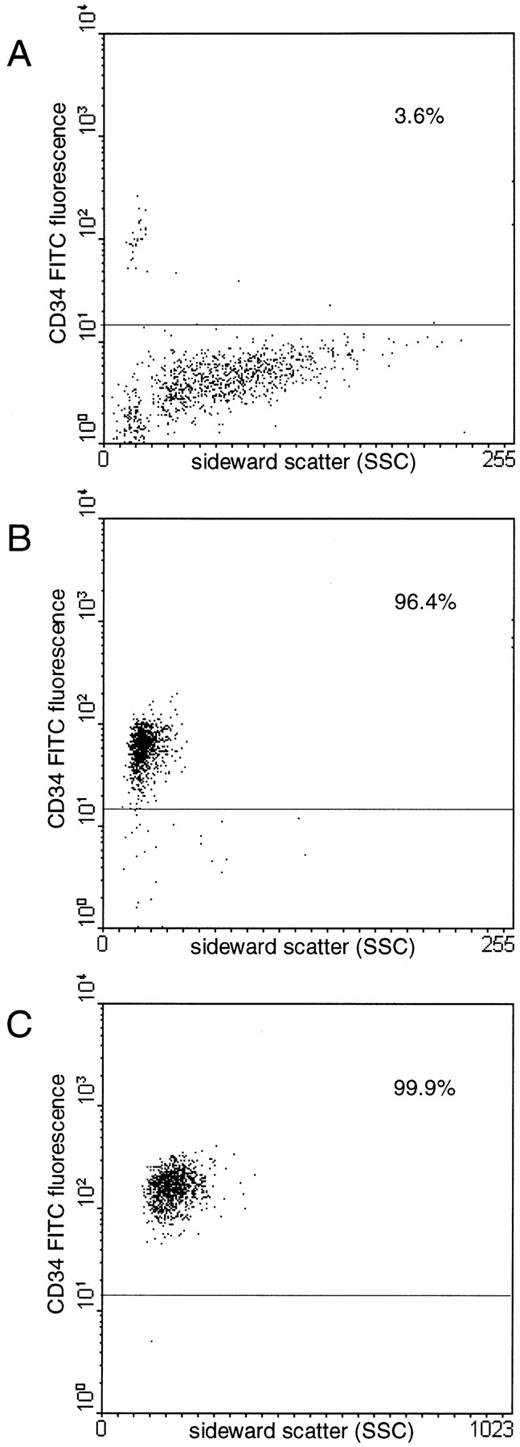
Purification of CD34+ hematopoietic progenitor cells.To obtain highly purified populations of hematopoietic progenitor cells for PCR analysis, blood-derived CD34+ cells from leukapheresis products were enriched using a two-step enrichment procedure. The median purity of CD34+ cells was 90.3% (range, 71.6% to 99.7%) after MiniMACSR separation and 99.8% (range, 99.7% to 99.9%) after FACS (Fig 1). The median numbers recovered were 5.2 × 106 CD34+ cells (range, 0.2 to 10.5 × 106), 1.8 × 106 CD34+/CD4+ cells (range, 0.1 to 4 × 106), and 2.4 × 106 CD34+/CD4− cells (range, 0.2 to 3.8 × 106).
Flow cytometric analyses of a representative CD34+ cell enrichment from a leukapheresis product (A) by immunomagnetic separation (B) and subsequent FACS (C). Proportions of CD34+ cells are indicated.
Flow cytometric analyses of a representative CD34+ cell enrichment from a leukapheresis product (A) by immunomagnetic separation (B) and subsequent FACS (C). Proportions of CD34+ cells are indicated.
Three-color immunofluorescence analysis of CD34+/CD4+ cells for HLA-DR expression.After immunomagnetic separation, the coexpression of CD4 was assessed on CD34+ cells from 14 samples. The median proportion of CD34 cells staining positive for CD4 was 34% (range, 11.3% to 58.5%). CD34+/CD4+ and CD34+/CD4− cells were gated and examined for HLA-DR expression using three-color fluorescence analysis (Table 1). The CD34+/CD4+ cell population had a lower HLA-DR expression in all samples except for one. The mean relative fluorescence intensity of HLA-DR on CD34+/CD4+ cells was 18 (range, 8.7 to 33.9), in comparison with 26 on CD34+/CD4− cells (range, 8.7 to 52.5; Table 1), whereas the mean relative fluorescence intensity of the isotype-identical antibody control was 3.7.
CD4 Expression on CD34+ Hematopoietic Progenitor Cells and HLA-DR Expression on CD34+/CD4+ and CD34+/CD4− Subpopulations
| Sample No. . | Proportion of . | Relative Mean Fluorescence . | |
|---|---|---|---|
| . | CD34+/CD4+ . | Intensity of HLA-DR . | |
| . | Cells (%) . | CD34+/CD4+ Cells . | CD34+/CD4− Cells . |
| 1 | 45.3 | 33.9 | 52.5 |
| 2 | 36.8 | 12.4 | 16.0 |
| 3 | 27.3 | 22.2 | 29.6 |
| 4 | 26.5 | 12.8 | 17.0 |
| 5 | 39.1 | 12.9 | 19.9 |
| 6 | 36.4 | 8.7 | 8.7 |
| 7 | 14.1 | 28.0 | 48.8 |
| 8 | 37 | 13.4 | 18.4 |
| Mean ± SD | 32.8 ± 9.1 | 18 ± 8.4 | 26.4 ± 15 |
| Sample No. . | Proportion of . | Relative Mean Fluorescence . | |
|---|---|---|---|
| . | CD34+/CD4+ . | Intensity of HLA-DR . | |
| . | Cells (%) . | CD34+/CD4+ Cells . | CD34+/CD4− Cells . |
| 1 | 45.3 | 33.9 | 52.5 |
| 2 | 36.8 | 12.4 | 16.0 |
| 3 | 27.3 | 22.2 | 29.6 |
| 4 | 26.5 | 12.8 | 17.0 |
| 5 | 39.1 | 12.9 | 19.9 |
| 6 | 36.4 | 8.7 | 8.7 |
| 7 | 14.1 | 28.0 | 48.8 |
| 8 | 37 | 13.4 | 18.4 |
| Mean ± SD | 32.8 ± 9.1 | 18 ± 8.4 | 26.4 ± 15 |
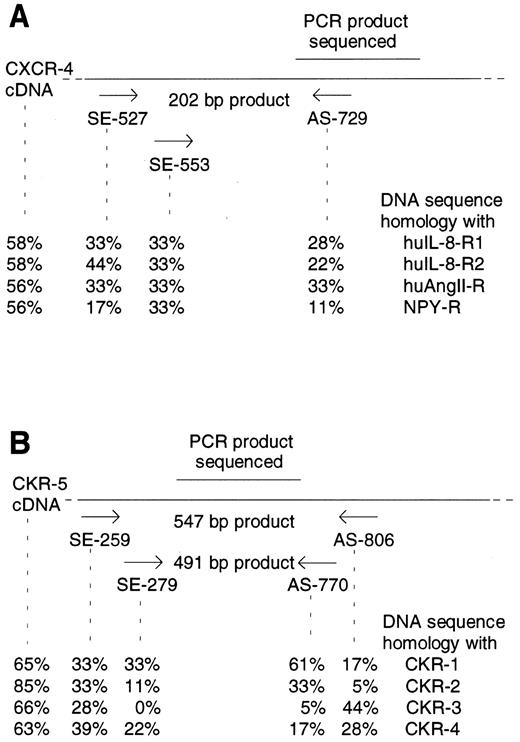
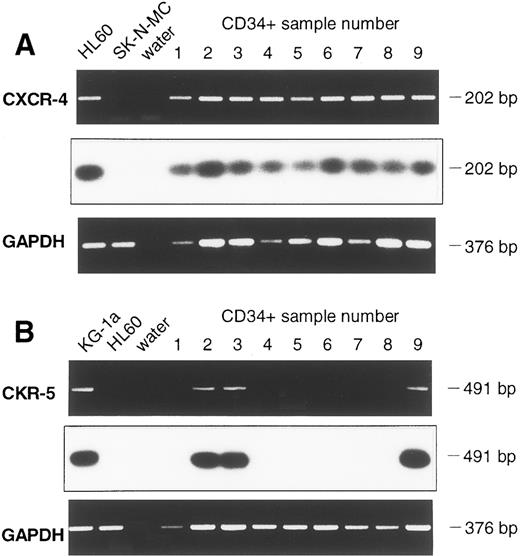
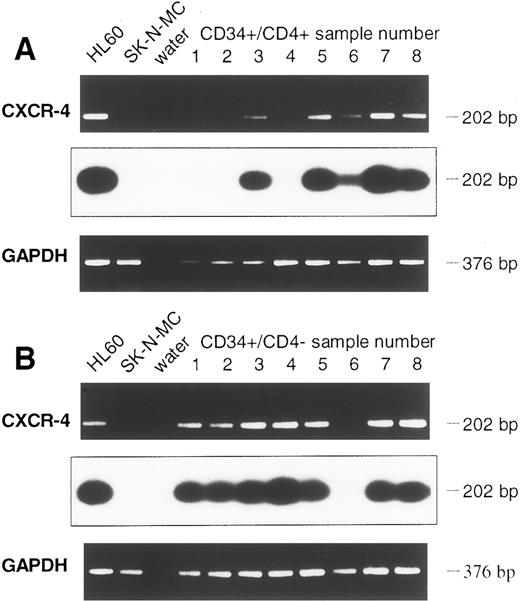
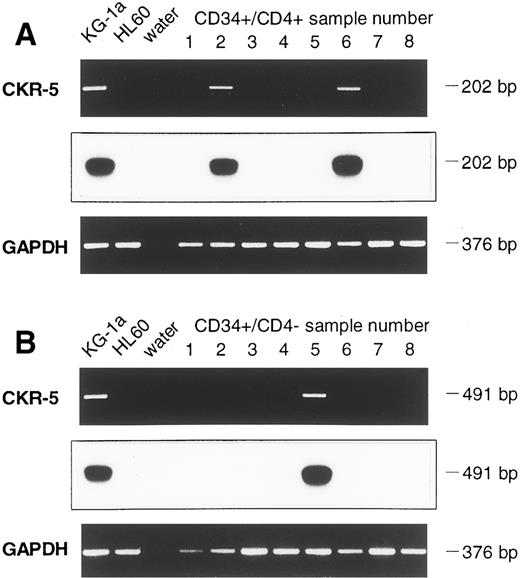
Detection of CXCR-4 and CKR5 mRNA in CD34+ and in CD34+/CD4+ cells.The sorted CD34+ cells were examined for the presence of mRNA of the HIV-1 coreceptors CXCR-4 and CKR-5. To prevent PCR amplification of related chemokine receptor cDNAs, CXCR-4 primers were chosen that had low sequence homology with the related cDNAs of human interleukin-8 (IL-8) receptor type 1 and 2 (huIL-8-R1 and huIL-8-R2), angiotensin II receptor (huAngII-R), and neuropeptide Y receptor (NPY-R; Fig 2A). Similarly, primers used for CKR-5 cDNA amplification also had low sequence homology with related chemokine receptors CKR-1, CKR-2, CKR-3, and CKR-4 cDNAs (Fig 2B). Different cell lines were used to establish the appropriate RT-PCR protocol. Confirming previous reports,16,17 19 the cell lines HL60 and CEM were positive for CXCR-4 mRNA, whereas SK-N-MC and U-87MG were negative. CXCR-4 mRNA was found in 9 samples of purified CD34+ cells (Fig 3A). Analyzing 8 further samples of CD34+ cells sorted for CD4 antigen, 5 of 8 CD34+/CD4+ samples and 7 of 8 CD34+/CD4− samples were positive for CXCR-4 mRNA (Fig 4A and B). Thus, in a total of 21 of 25 samples examined (84%) expression of CXCR-4 mRNA was observed.
Position of CXCR-4 (A) and CKR-5 (B) specific primers and sequence homologies with related chemokine receptor cDNAs. Expected lengths of PCR products are indicated. DNA sequence homologies of PCR primers and of CXCR-4 and CKR-5 cDNA with the related chemokine receptor cDNAs were determined using HUSAR. EMBL data base accession numbers: CXCR-4 (X71635), huIL-8-R1 (L19591), huIL-8-R2 (M74240), huAngII-R (M93394), NPY-R (M88461), CKR-1 (L09230), CKR-2 (U03882), CKR-3 (U28694), CKR-4 (X85740), and CKR-5 (X91492).
Position of CXCR-4 (A) and CKR-5 (B) specific primers and sequence homologies with related chemokine receptor cDNAs. Expected lengths of PCR products are indicated. DNA sequence homologies of PCR primers and of CXCR-4 and CKR-5 cDNA with the related chemokine receptor cDNAs were determined using HUSAR. EMBL data base accession numbers: CXCR-4 (X71635), huIL-8-R1 (L19591), huIL-8-R2 (M74240), huAngII-R (M93394), NPY-R (M88461), CKR-1 (L09230), CKR-2 (U03882), CKR-3 (U28694), CKR-4 (X85740), and CKR-5 (X91492).
Detection of CXCR-4 (A) and CKR-5 (B) mRNA in samples of CD34+ cells of 9 leukapheresis products using RT-PCR. Chemokine mRNA specific PCR, Southern Blot hybridization, and GAPDH-PCR are shown one below another. For CXCR-4 mRNA, the cell line HL60 served as a positive control and SK-N-MC and water as negative controls. CKR-5 mRNA analysis of cell line KG-1a was a positive control, whereas HL60 and water were negative controls.
Detection of CXCR-4 (A) and CKR-5 (B) mRNA in samples of CD34+ cells of 9 leukapheresis products using RT-PCR. Chemokine mRNA specific PCR, Southern Blot hybridization, and GAPDH-PCR are shown one below another. For CXCR-4 mRNA, the cell line HL60 served as a positive control and SK-N-MC and water as negative controls. CKR-5 mRNA analysis of cell line KG-1a was a positive control, whereas HL60 and water were negative controls.
Detection of CXCR-4 mRNA in CD34+ cell samples of eight further leukapheresis products, sorted for CD34+/CD4+ (A) and CD34+/CD4− (B) cells. Controls and Southern blot hybridization are corresponding to Fig 3.
Detection of CXCR-4 mRNA in CD34+ cell samples of eight further leukapheresis products, sorted for CD34+/CD4+ (A) and CD34+/CD4− (B) cells. Controls and Southern blot hybridization are corresponding to Fig 3.
Assessing cell lines for CKR-5 mRNA, the cell line KG1a was positive, whereas HUT-78, HL60, and K562 were negative, which is in line with previous reports.21,22 25 Whereas CKR-5 mRNA was found in KG1a in a one-step PCR, only 3 of 9 CD34+ cell samples were positive for CKR-5 mRNA, even though a two-step (nested) PCR was used (Fig 3B). Similar results were obtained when 8 further samples of CD34+ cells were assessed after FACS sorting for CD4. Using the nested RT-PCR protocol, CKR-5 mRNA was present in 2 of 8 CD34+/CD4+ cell samples and in 1 of 8 CD34+/CD4− cell samples (Fig 5A and B). As a result, a total of 6 of 25 samples assessed (24%) contained CKR-5 mRNA.
Detection of CKR-5 mRNA in CD34+/CD4+ (A) and in CD34+/CD4− (B) cell samples (B). Controls and Southern blot hybridization are corresponding to Fig 3.
Detection of CKR-5 mRNA in CD34+/CD4+ (A) and in CD34+/CD4− (B) cell samples (B). Controls and Southern blot hybridization are corresponding to Fig 3.
All PCRs performed, except the water controls, were positive for GAPDH, showing the presence of amplifyable cDNA (Figs 3, 4, and 5). Omitting reverse transcription, all samples were negative in CXCR-4, CKR-5, and GAPDH RT-PCR, demonstrating the absence of contaminating genomic DNA (data not shown).
Hybridization of PCR products using recombinant CXCR-4 and CKR-5 DNA.To determine the specificity of the PCR products and to exclude that the negative PCRs were below the detection level of the agarose gel electrophoresis, all PCR products were analyzed by Southern hybridization. The hybridization signals of all CXCR-4 and CKR-5 PCR products were in accordance with the results obtained from ethidiumbromide staining of agarose gels (Figs 3, 4, and 5). GAPDH and CC-CKR5 PCR products did not hybridize with the recombinant CXCR-4 DNA probe, and GAPDH or CXCR-4 PCR products did not hybridize with the recombinant CKR-5 DNA probe (data not shown).
Sequence analysis of CXCR-4 and CKR-5 PCR products.To confirm that CXCR-4 and CKR-5 mRNA but no related sequences were amplified in the PCR reactions, cycle sequencing of PCR products was performed. The sequences of two CXCR-4 PCR products were determined for 83 nucleotides, which were identical with the CXCR-4 cDNA sequence from position 709 to 791 (EMBL data bank accession no. X71635). Similarly, the sequence of the 111 nucleotides assessed for two CKR-5 PCR products was identical with the CKR-5 cDNA sequence from position 411 to 521 (EMBL data bank accession no. X91492).
DISCUSSION
In this report, we show that a subset of peripheral blood-derived CD34+ cells are potential target cells for HIV-1 infection. This can be concluded from the presence of CXCR-4 mRNA in the majority of CD34+ cell samples. CXCR-4 mRNA was present in both subsets, CD34+/CD4+ cells as well as CD34+/CD4− cells, suggesting that expression of CXCR-4 is not related to CD4 expression. The latter antigen was predominantly found on CD34+ cells with a relative low fluorescence intensity for HLA-DR, which is compatible with a more immature subset of hematopoietic progenitor cells.31 This would imply that CD34+ cells at an earlier stage of development are particularly prone for an HIV infection. On the other hand, in semisolid culture assays, HIV-1 was only found in granulocyte-macrophage (colony-forming unit–granulocyte-macrophage [CFU-GM]) and erythroid (burst-forming unit-erythroid [BFU-E]), but not from pluripotent progenitor cells (colony-forming unit – granulocyte-erythroid-monocyte-megakaryocyte [CFUGEMM]).13
The CXCR-4 chemokine receptor for which we found mRNA in the CD34+ cell population is a transmembrane protein that serves T-cell–tropic HIV-1 strains as a coreceptor for infection.16 Indeed, CD34+ bone marrow cells could be infected with T-cell–tropic HIV-1 strains in vitro,11-13 suggesting CXCR-4 expression in a functional state and sufficient density on CD34+ cells. Transcription of mRNA as assessed in this work must not necessarily mean expression of protein. Still, the presence of functional CXCR-4 on the surface of CD34+ cells is very likely, because stromal derived factor-1 (SDF-1) is a natural ligand for this chemokine receptor.20 SDF-1, produced by murine bone marrow stromal cells,20 is the first chemoattractant reported for human CD34+ hematopoietic progenitor cells.32,33 This view is supported by in vivo studies showing that mice genetically deficient for SDF-1 have myelopoiesis in the fetal liver but not in the bone marrow.34 CXCR-4 on CD34+ hematopoietic progenitor cells may therefore be involved in the homing of circulating progenitor cells to the bone marrow stroma by means of SDF-1 binding.
CKR-5 serves macrophage-tropic HIV-1 strains as a coreceptor for infection.21 Accordingly, CKR-5 ligands RANTES (regulated on activation, normal T-cell expressed and secreted), macrophage inflammatory protein-1α (MIP-1α), and MIP-1β are secreted by CD8+ T cells and inhibit the infection with macrophage-tropic but not T-cell–tropic HIV-1 isolates.22,24,25 35 In our experiments, CKR-5 was only detected in 2 of 8 samples of CD34+/CD4+ hematopoietic progenitor cells, even though a nested PCR was used. This finding either reflects a low incidence of CKR-5 transcription or contamination of the enriched CD34+ cells by minute numbers of monocytes or macrophages. Because positivity for CKR-5 mRNA did not correlate with the amount of RNA assessed, low transcription of CKR-5 mRNA in the CD34 cells of positive samples is the more likely explanation for our data.
Most virus isolates from HIV-1 patients shortly after seroconversion and during the asymptomatic stage are macrophage-tropic, whereas T-cell–tropic isolates are more frequently observed in patients with AIDS.36 T-cell–tropic HIV-1 strains therefore herald a more aggressive course of the disease, because they induce the formation of syncytia in CD4+ cells and replicate in peripheral blood mononuclear cells at a higher rate than macrophage-tropic strains.37 Phenotype switch of HIV-1 from macrophage- to T-cell–tropic virus strains could result from HIV-1 Env mutation during the course of the disease,38 which is supported by the finding of a primary HIV-1 isolate that uses both CKR-5 and CXCR-4 as entry cofactors.23 Another explanation for phenotype switch may be a decreasing B-cell response, keeping in check T-cell–tropic HIV-1 species during the early stages of the disease but not in AIDS.39
From our data we conclude that macrophage-tropic HIV-1 strains as observed during the asymptomatic stage of the infection would not infect CD34+ hematopoietic progenitor cells because of the lack or low expression of CKR-5. This conclusion is compatible with published data that CD34+ cells are not a major reservoir of HIV-1.7,8,40,41 Later in the course of the disease, HIV-1 may develop a T-cell–tropic phenotype with the ability to infect a subset of hematopoietic progenitor cells expressing CD4 and CXCR-4. This model could explain the finding that HIV-1 strains isolated from CD34+ cells of 10 patients with AIDS were in all cases T-cell–tropic.5
ACKNOWLEDGMENT
The authors thank Michael Kirsch, Margit Pförsich, and Klaus Hexel for excellent technical assistance and Georg Sczakiel for continuous support.
Address reprint requests to Rainer Haas, MD, Medizinische Klinik und Poliklinik V der Universität Heidelberg, Hospitalstraβe 3, 69115 Heidelberg, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal