Abstract
Our previous work in patients undergoing autologous transplant for multiple myeloma (MM) or breast cancer (BC) has shown that retroviral transduction of adult CD34+ cells for 72 hours in the presence of interleukin-3 (IL-3), IL-6, and stem cell factor (SCF ) resulted in .01% to 1% long-term marking of peripheral blood and marrow cells (Blood 85:3948, 1995). In this study we compare these previous studies to transduction with no added growth factors, previously shown to result in higher levels of marking in children (Lancet 342:1134, 1993) or transduction in the presence of an autologous stromal layer. Peripheral blood (PB) mononuclear cells were collected via apheresis after high-dose cyclophosphamide and granulocyte colony-stimulating factor. Bone marrow (BM) was also harvested in all patients. One third of both BM and PB collections were enriched for CD34+ cells and transduced with one of two marking vectors containing the neomycin-resistance gene to distinguish cells originating from BM and PB posttransplantation. Cells from 3 MM and 2 BC patients were transduced without growth factors for 6 hours and cells from 2 MM and 2 BC patients were transduced in the presence of autologous marrow stroma. Immediately posttransduction, the percentage of Neo-resistant PB and BM progenitors (colony-forming units) were: 0% to 19% in the 6-hour no growth factor group and 0% to 36% in the autologous stroma group. After conditioning therapy, both transduced and untransduced PB and BM fractions were infused into the patients. Semi-quantitative nested DNA polymerase chain reaction was performed on total, mononuclear, and granulocyte fractions of PB and BM at 1, 3, 6, 9, 12, and 18 months. Poor marking has been observed in both groups, with no consistently positive patients. These results compare unfavorably with our prior experience using growth factors during transduction. Further optimization of transduction conditions and vectors needs to be developed to improve transduction efficiency of adult human repopulating hematopoietic cells.
THE HEMATOPOIETIC stem cell is an obvious target for gene transfer because it is defined by its potential to reconstitute the hematopoietic and immune systems.1,2 Retroviral gene transfer is the best characterized and most developed vector system and is currently under the widest clinical study.3 Although efficient and reproducible retroviral gene transfer to a high percentage of long-term repopulating cells has been shown in rodents, only limited success has been achieved in larger animals, including humans.1 Although murine bone marrow (BM) cultured with retrovirus in the presence of interleukin-3 (IL-3), IL-6, and stem cell factor (SCF ) has provided efficient gene transfer to a significant percentage of hematopoietic stem cells, previous work from our group has shown only minimal levels of gene transfer in circulating progenitors of human long-term repopulating cells when CD34+ cells were transduced using a similar protocol.4 Higher levels of gene transfer were reported in children recovering from high-dose chemotherapy when their marrow cells were simply exposed to retroviral vector supernatant for 6 hours without the inclusion of exogenous growth factors.5 Stromal cells or stromal matrix molecules have been shown to substitute for or enhance the effects of exogenous growth factors during transduction of human hematopoietic progenitors and improve gene transfer into primitive cells in in vitro testing and in animal models.6-8 In this study we compared the 6-hour transduction procedure without growth factors in adults to the use of autologous marrow stromal during transduction as a means of improving efficiency of gene transfer into primitive repopulating cells. We report here the results in nine patients observed for at least 6 months after autologous transplantation of retrovirally transduced peripheral blood (PB) and BM CD34-enriched cells.
MATERIALS AND METHODS
Clinical procedures.After informed consent, patients were enrolled in ongoing Institutional Review Board–approved autologous transplantation protocols for chemotherapy-responsive multiple myeloma (MM) or breast cancer (BC) at the Warren Grant Magnuson Clinical Center of the National Institutes of Health (Bethesda, MD). The overall transplant procedure has been published previously4,9 and differs here only in the retroviral transduction conditions summarized below. Figure 1 shows the clinical and laboratory procedures. Characteristics of the nine patients enrolled into the gene marking protocol over a 2-year period are summarized in Table 1. Our protocol design has been published previously4 and differs here only in the transduction conditions used. As in our previous clinical protocol of retroviral transduction in the presence of IL-3, IL-6, and SCF, CD34-enriched fractions of the PB and BM were used for transduction. Some of the MM patients had their entire BM and PB grafts CD34-enriched, not just those fractions used for transduction. These patients are identified in Table 1.
Schema of gene-marking protocol. ICE: ifosphamide, carboplatin, etoposide; MM: multiple myeloma patients; BC: breast cancer patients.
Schema of gene-marking protocol. ICE: ifosphamide, carboplatin, etoposide; MM: multiple myeloma patients; BC: breast cancer patients.
Patient Characteristics and Transduction Data
| Patient . | Age/Sex . | Cell Source . | Vector . | CD34 Cells . | CFU-C Transduction Efficiency %* . | Transplant Date . | Status . |
|---|---|---|---|---|---|---|---|
| . | . | . | . | Infused/kg . | . | . | . |
| . | . | . | . | (millions) . | . | . | . |
| 6H | 57/M | PB | G1Na | 1.46 | 19 | 6/20/95 | Alive/CR |
| MM 1 | PB | None | 9.05 | ||||
| BM | LNL6 | 0.20 | 8 | ||||
| BM | None | 0.81 | |||||
| 6H | 44/M | PB | G1Na | 0.08 | >1 | 4/7/94 | Alive/PR |
| MM 2 | PB | None | 0.80 | ||||
| BM | LNL6 | 0.75 | ND | ||||
| BM | None | 0.75 | |||||
| 6H | 54/M | PB | G1Na | 0.01 | 1 | 11/18/94 | Alive/PR |
| MM 3 | PB | None | 1.63 | ||||
| BM | LNL6 | 0.07 | 0 | ||||
| BM | None | 0.02 | |||||
| 6H | 45/F | PB | LNL6 | 4.61 | >0 | 10/11/94 | Alive/CR |
| BC 4 | PB | None | 19.29 | ||||
| BM | G1Na | 0.18 | 2 | ||||
| BM | None | 4.00 | |||||
| 6H | 53/F | PB | LNL6 | 0.80 | ND | 10/12/95 | Alive/PD Day 111 |
| BC 5 | PB | None | 3.40 | ||||
| BM | G1Na | 0.11 | ND | ||||
| BM | None | 1.00 | |||||
| ST | 46/M | PB | LNL6 | 0.07 | 20 | 6/24/94 | Alive/PR |
| MM 1 | PB | None | 1.03 | ||||
| BM | G1Na | 0.00 | 2 | ||||
| BM | None | 0.36 | |||||
| ST | 48/M | PB | G1Na | 1.12 | 0 | 6/10/94 | Alive/PR |
| MM 2 | PB | None | 27.55 | ||||
| BM | LNL6 | 0.15 | 10 | ||||
| BM | None | 1.69 | |||||
| ST | 49/F | PB | LNL6 | 0.09 | ND | 2/29/96 | Alive/PR |
| BC 3 | PB | None | 0.72 | ||||
| BM | G1Na | 0.18 | 30 | ||||
| BM | None | 6.63 | |||||
| ST | 36/F | PB | G1Na | 0.32 | 36 | 3/12/96 | Alive/PD Day 123 |
| BC 4 | PB | None | 1.10 | ||||
| BM | LNL6 | 0.37 | ND | ||||
| BM | None | 4.31 |
| Patient . | Age/Sex . | Cell Source . | Vector . | CD34 Cells . | CFU-C Transduction Efficiency %* . | Transplant Date . | Status . |
|---|---|---|---|---|---|---|---|
| . | . | . | . | Infused/kg . | . | . | . |
| . | . | . | . | (millions) . | . | . | . |
| 6H | 57/M | PB | G1Na | 1.46 | 19 | 6/20/95 | Alive/CR |
| MM 1 | PB | None | 9.05 | ||||
| BM | LNL6 | 0.20 | 8 | ||||
| BM | None | 0.81 | |||||
| 6H | 44/M | PB | G1Na | 0.08 | >1 | 4/7/94 | Alive/PR |
| MM 2 | PB | None | 0.80 | ||||
| BM | LNL6 | 0.75 | ND | ||||
| BM | None | 0.75 | |||||
| 6H | 54/M | PB | G1Na | 0.01 | 1 | 11/18/94 | Alive/PR |
| MM 3 | PB | None | 1.63 | ||||
| BM | LNL6 | 0.07 | 0 | ||||
| BM | None | 0.02 | |||||
| 6H | 45/F | PB | LNL6 | 4.61 | >0 | 10/11/94 | Alive/CR |
| BC 4 | PB | None | 19.29 | ||||
| BM | G1Na | 0.18 | 2 | ||||
| BM | None | 4.00 | |||||
| 6H | 53/F | PB | LNL6 | 0.80 | ND | 10/12/95 | Alive/PD Day 111 |
| BC 5 | PB | None | 3.40 | ||||
| BM | G1Na | 0.11 | ND | ||||
| BM | None | 1.00 | |||||
| ST | 46/M | PB | LNL6 | 0.07 | 20 | 6/24/94 | Alive/PR |
| MM 1 | PB | None | 1.03 | ||||
| BM | G1Na | 0.00 | 2 | ||||
| BM | None | 0.36 | |||||
| ST | 48/M | PB | G1Na | 1.12 | 0 | 6/10/94 | Alive/PR |
| MM 2 | PB | None | 27.55 | ||||
| BM | LNL6 | 0.15 | 10 | ||||
| BM | None | 1.69 | |||||
| ST | 49/F | PB | LNL6 | 0.09 | ND | 2/29/96 | Alive/PR |
| BC 3 | PB | None | 0.72 | ||||
| BM | G1Na | 0.18 | 30 | ||||
| BM | None | 6.63 | |||||
| ST | 36/F | PB | G1Na | 0.32 | 36 | 3/12/96 | Alive/PD Day 123 |
| BC 4 | PB | None | 1.10 | ||||
| BM | LNL6 | 0.37 | ND | ||||
| BM | None | 4.31 |
Bold type indicates that all mononuclear cells, not just CD34+ cells, were infused.
Abbreviations: MM, multiple myeloma; BC, breast cancer; PR, partial remission; CR, complete remission; PD, progressive disease; CFU-C, colony-forming unit cell.
% Neomycin resistant CFU-C assayed at the end of transduction.
Viral vectors.Two retroviral vectors, G1Na.40 and LNL6, carrying an identical bacterial phosphotransferase gene conveying G418 neomycin resistance (Neo) were used. Clinical grade supernatants obtained from producer cell lines grown to confluence in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum were obtained from Genetic Therapy Inc (Gaithersburg, MD). All supernatants were shown to be free of replication-competent helper virus using the direct S + L- complementation focus assay, and all lots were also tested by coculture amplification on the mus dunni cell line.10 The vector titer of each aliquot of supernatant used ranged from 4.2 × 105 to 2.1 × 106 biologically active particles/mL, which was the same titer range as our previous study.4
Transduction with retroviral vectors.Two distinct marking vectors carrying the Neomycin resistance gene (Neo) were used to distinguish the PB from the BM grafts. BM and PB from five patients were transduced for 6 hours without the addition of other growth factors, herein referred to as ‘6H’ conditions or patients, and BM and PB from four patients were transduced for 72 hours in the presence of autologous stroma, herein referred to as ‘ST’ conditions or patients. For patients entered in the 6H protocol, CD34-enriched BM or PB cells were transduced in clinical-grade supernatants at a density of 1 to 2 × 105/mL for 6 hours at 37°C in 5% CO2 . For ST patients, CD34-enriched cells from PB or BM at a density of 1 to 2 × 105/mL were cocultured with 70% to 100% confluent autologous marrow stromal layers in the presence of vector supernatant. Supernatant and nonadherent cells were collected and centrifuged every 24 hours. The nonadherent cells were resuspended in fresh vector supernatant and returned to the same stromal culture flask. The total transduction period was 72 hours. If LNL6 was used for the PB, G1Na.40 was used for the BM, and vice versa. Both 6H and ST transduction cultures were supplemented with 4 μg/mL protamine sulfate (Elkins-Sinn, Cherry Hill, NJ) and 50 mg/mL gentamycin sulfate (GIBCO, Gaithersburg, MD). At the end of the culture period nonadherent cells were collected and both stromal and hematopoietic cells adhering to the culture flask were dislodged by exposure to 0.25% trypsin (GIBCO) for 5 minutes. All cells were washed three times and counted. In the ST arm, small round hematopoietic cells were counted separately from the much larger stromal cells. Aliquots were removed to assess viability, CD34 expression, colony-forming unit cell number, G418 resistance, and transduction efficiency as assessed by polymerase chain reaction (PCR) for the Neo gene. Cells were then cryopreserved in a controlled rate freezer.
Autologous stromal cells.At least 3 to 4 weeks before receiving cyclophosphamide, approximately 10 mL of marrow aspirate collected from the posterior superior iliac crest of each ST patient was purified for mononuclear cells on a Ficoll-hypaque density gradient (Lymphocyte Separation Medium; Organon Teknika, Durham, NC). Washed cells were plated at a density of 1 to 2 × 106 cells/mL in Dulbecco's modified Eagle's medium supplemented with 20% fetal calf serum, 2 mmol/L L-glutamine, and 10 μg/mL gentamycin and incubated at 37°C in 5% CO2 . Medium was changed at 24 hours and every 3 to 4 days until the adherent stroma was confluent. Cells were then exposed to 0.25% trypsin (GIBCO) for 5 minutes, washed, and expanded into T-162–cm2 flasks at 3 to 4 × 105 cells per flask. Repeated rounds of expansion, typically over 2 to 4 weeks, eventually produced enough 70% to 100% confluent T-162 — cm2 flasks to transduce CD34-enriched cells at a concentration of 1 × 105 cells/mL with a maximum of 150 mL total medium per flask. Flasks were exposed to 1,000 cGy irradiation on the day of transduction.
Posttransduction analysis.CFU plating and analysis and cell separations on posttransplant sample and DNA extractions were done as described previously.4 The PCR procedures used to analyze patient blood and BM samples for the Neo gene and the amphotrophic envelope gene have been outlined in detail previously.4
RESULTS
Protocol design, CD34 enrichment results, and clinical outcomes.In all patients, one third of the harvested BM or one of three mobilized PB collections were CD34-selected and transduced with either LNL6 or G1Na.40 marking vectors. For the five 6H patients, CD34-enrichment of PB resulted in postimmunoabsorption fractions that averaged 70.8% (SD 14.5) CD34+ cells. After transduction, the CD34 percentage decreased to 65% (SD 19.4). Total and CD34+ cell counts also decreased over the 6 hours of transduction by a mean of 37% (SD 16) and 50% (SD 14), respectively. After immunoabsorption the BM fractions were 67.6% (SD 13.9) CD34+. After transduction, the CD34+ percentage decreased to 58.7% (SD 26). Total and CD34+ cell counts decreased in the BM as well by a mean of 43% (SD 24) and 57% (SD 30).
For the four ST patients, CD34-enrichment of PB resulted in postimmunoabsorption fractions that averaged 50% (SD 29) CD34+ cells. After 72 hours of transduction, the CD34 percentage decreased to 38% (SD 33). Total and CD34+ cell counts also decreased over the 72 hours of transduction by a mean of 30% (SD 17) and 56% (SD 10), respectively. After immunoabsorption the BM fractions were 67% (SD 15) CD34+. After transduction, the percentage of CD34+ cells decreased to 53% (SD 8.5). Total and CD34+ cell counts fell in the BM as well by a mean of 20% (SD 16) and 38% (SD 23).
Table 1 shows the total number of transduced and nontransduced CD34+ PB and BM cells infused into each patient. Engraftment in the two 6H patients who received only CD34-selected BM cells (6H-MM1 and 6H-MM3) did not differ from the rest of the group. There was wide variability in the total and CD34+ cells/kg infused and in the ratio of transduced to untransduced cells infused. The wide variability is likely caused by differences in individual hematopoietic reserve resulting from prior chemotherapy, and perhaps differential sensitivity of the cells to the transduction conditions. CD3+ T cells were depleted 2.5 to 4 logs in both BM and PB grafts by the CD34-enrichment procedure.11
Patients receiving transduced CD34+ cells engrafted on schedule compared with patients in our previous gene-marking protocol and identical clinical protocols without gene marking, ie, neutrophil counts of 500/μL were achieved by a mean day of 11, and platelets reaching 20,000/μL by a mean of day 15 posttransplantation. No toxicity attributable to the marking procedures was seen, such as acute reactions during infusion of transduced cells or stroma or unusual infections. Because of concern over adverse consequences of replication-competent helper virus generation, posttransplantation PB mononuclear cell DNA samples from each patient were tested for recombinant helper virus as described previously.4 No samples have been positive, with a sensitivity of detection of 1 genome per 10,000 cells (data not shown).
In vitro assessment of transduction efficiency.Aliquots of transduced PB and BM cells from each patient were assayed by two different methods. Cells were plated in methylcellulose-containing media with and without G418 at 1.2 μg/mL (active), and the percentage of CFU-C (colony-forming unit cell) resistant to G418 was calculated. Table 1 gives the individual values for each transduction. Overall marking efficiency was poor, with great variability between patients and between PB and BM fractions for each patient. There was a significantly greater mean transduction efficiency in the CD34-enriched cells transduced on stroma compared with the 6H group (19.6 v 7.5, respectively; P = .01). Cell pellets from the end of transduction were also assessed by semiquantitative PCR for the frequency of the transferred Neo gene; this assay provided similar estimates of gene transduction efficiency compared with CFU-C Neo percentages (data not shown).
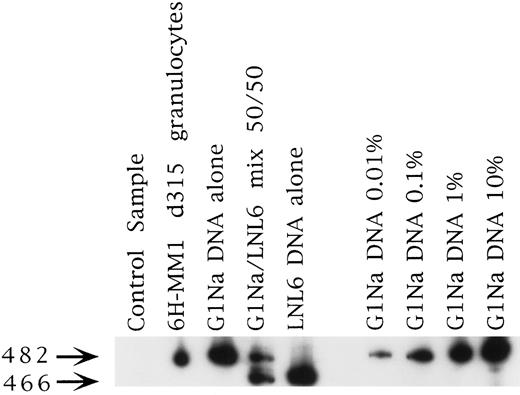
Posttransplantation analysis for the marker gene.DNA samples prepared from PB, granulocyte and total mononuclear PB fractions, and BM were analyzed by semi-quantitative PCR for the Neo gene at the time of engraftment and then every 3 months posttransplantation for up to 24 months. Representative PCR results are shown in Fig 2. Each set of patient samples collected had a normal donor PB sample processed in parallel to control for any contamination that may have been introduced during processing. Patient samples were only scored as positive if the control DNA samples were negative. No control samples were found to be positive.
Representative PCR sample from patient 6H-MM1. This was the longest marking observed in any patient at 315 days. The signal strength is about 0.1% and is from the fraction marked with the G1Na vector, which was used to mark the PB fraction in this case (see Table 1).
Representative PCR sample from patient 6H-MM1. This was the longest marking observed in any patient at 315 days. The signal strength is about 0.1% and is from the fraction marked with the G1Na vector, which was used to mark the PB fraction in this case (see Table 1).
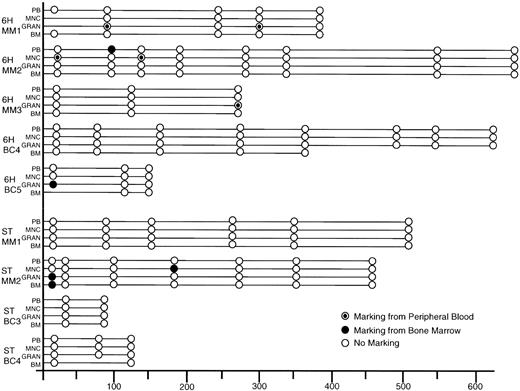
Figure 3 summarizes the Neo marking data over time posttransplantation in all patients. Only two 6H patients and one ST patient had detectable gene marked cells immediately posttransplant. Patient 6H-MM2 showed mononuclear cell marking from the PB graft fraction at 0.1% and patient 6H-BC5 showed granulocyte marking originating from the BM at 0.1%. Patient ST-MM2 had signals in his granulocyte and BM cells derived from the BM graft at 0.1% and 0.01%, respectively. Early posttransplantation analysis in our previous cohort of patients whose cells were transduced in the presence of growth factors IL-3, IL-6, and SCF for 72 hours had shown Neo marking in all patients at levels ranging from 0.01% to 1%.
Summary of PCR results on all patient samples post-transplantation. Samples were scored as negative if β-actin PCR of the sample gave a signal consistent with amplification of 500 ng of DNA. Samples were scored as positive only if both reagent and concurrently extracted normal PB controls were negative for the Neo signal. (○), Negative and (•), positive for vector used to transduce the BM, and (⊙), positive for the vector used to transduce the PB. MNC, mononuclear fraction of PB; GRAN, granulocyte fraction of PB.
Summary of PCR results on all patient samples post-transplantation. Samples were scored as negative if β-actin PCR of the sample gave a signal consistent with amplification of 500 ng of DNA. Samples were scored as positive only if both reagent and concurrently extracted normal PB controls were negative for the Neo signal. (○), Negative and (•), positive for vector used to transduce the BM, and (⊙), positive for the vector used to transduce the PB. MNC, mononuclear fraction of PB; GRAN, granulocyte fraction of PB.
For patient ST-MM2, the one ST patient with gene-marked cells posttransplantation, the marked cells were detected 186 days posttransplantation in 0.1% of the mononuclear PB fraction derived from the transduced BM graft. This is consistent with the higher transduction efficiency of progenitors seen in this transduced BM (10%) as compared with PB (0%). Furthermore, this patient had higher transduced CD34-enriched cell numbers infused as compared with the other ST patients.
6H-MM1 had marked granulocytes detected 315 days posttransplantation at a level of 0.1%. The short circulation and survival time of granulocytes means that the positive PCR signal was not simply caused by prolonged survival of transduced terminally differentiated cells, but rather by production of daughter cells from previously quiescent progenitors. Patients 6H-MM1 and 6H-BC5 also showed early marking in their granulocyte fractions from the PB and BM grafts, respectively. No later marking could be detected in these patients, suggesting that only progenitors had been marked in these patients or that the level of marked cells had fallen below the detection limits of our PCR assay.
The fact that low-level marking was observed in 4 of 5 6H patients versus 1 of 4 ST patients suggests that the 6H condition was more favorable for gene transfer. In contrast to our previous study, none of the 6H or ST patients showed consistent marking over time or had any marked cells detected greater than 1 year post transplantation.
Only patient ST-BC4 with relapsing disease posttransplantation has been evaluated for gene marking of tumor cells. Nested Neo PCR of tumor cell DNA extracted from malignant pleural effusion cells on day 123 was negative. Neither BC nor MM cells express CD34 antigen and the CD34-enrichment process has been shown to serve as a 2- to 4-log purge of myeloma and BC tumor cells, so very few tumor cells would have been exposed to retroviral vector during transduction in any case.12 13
DISCUSSION
The overall efficiency of retroviral gene transfer in this clinical trial was not better than that seen in our previous cohort of patients whose cells were transduced in the presence of growth factors, and is thus unlikely to be useful for therapeutic applications. Use of CD34-enriched target cells allowed us to preserve an optimal multiplicity of infection, ie, at least 10 infectious viral particles/cell. We and others have previously shown CD34-enriched BM and PB cells are equal or better targets for gene transfer as compared with whole mononuclear cells.14 15 No assessment of the relative potential of BM versus PB CD34-enriched cells to serve as targets for retroviral gene transfer under 6H and ST conditions could be done because of the generally poor level of gene transfer.
The gene transfer efficiency observed in our 6H patients was less than that observed for children whose cells were transduced under the same conditions with identical vectors at St Jude Children's Research Hospital (Memphis, TN).5 Levels of marked BM CFU-C there averaged 5% at 12 to 18 months posttransplantation and have continued to be detected and expressed for up to 3 years in BM.16 The differences between our two protocols may explain the marking disparity and include patient age, timing of marrow collection, assay for gene transfer, cell type transduced, and disease treated (acute myeloid leukemia [AML] and neuroblastoma v MM and BC). The younger age of the St Jude patients (range, 2 to 19 years) and the prompt collection of autologous marrow after high-dose induction chemotherapy for relapsed tumor may have provided a higher number of primitive hematopoietic cells in cycle and susceptible to retroviral gene transfer.17 In contrast, our patients were middle-aged adults heavily pretreated with multiple cycles of moderate-dose myelosuppressive therapy and may thus have sustained permanent stem cell depletion or damage. Alternatively, different methods for determining marking efficiency posttransplant may have skewed the comparison. We chose a strategy of nested PCR on total DNA from BM and PB fractions, whereas the marking efficiency of the St Jude patients was evaluated by DNA PCR on colonies of Neo-resistant progenitor colonies grown out at various time points posttransplant.
Discrepancies between BM Neo-resistant colony and total DNA PCR on mature circulating cells have been noted in other reports.18-20 This observation has not been systematically evaluated thus far. Immune responses against intracellular marker genes such as the hygromycin resistance gene have been reported in trials using mature lymphocytes as target cells.21,22 It is possible that the Neo gene product is expressed more highly in mature circulating cells, leading to immune destruction despite persistence of marrow progenitors. However, in our study we did not find detectable persistence of transduced marrow progenitors. It is also possible that the Neo gene product has direct toxicity to some hematopoietic cell types.23
Whether our use of a CD34-enriched cell population might have decreased gene transfer efficiency when compared with the total mononuclear population used at St Jude is unclear. Studies suggest that the CD34+ population is equally susceptible to gene transfer when compared with the total mononuclear population.14,15 However, interactions between accessory cells might be important in promoting stem cell survival and proliferative potential without terminal differentiation. Our previous group of patients that had cells transduced in the presence of growth factors had an expansion in their total and CD34 cell numbers (Table 2); some of these cells may have served an accessory function. Evidence against this hypothesis comes from the poor gene transfer seen in a different cohort of adults with leukemia in remission who underwent autologous transplant with total BM mononuclear cells transduced with the same G1Na vector we used for 4 hours without growth factors.24 Under both ST and 6H conditions, total and CD34 cell numbers decreased markedly. Hence, either primitive cell cycling or accessory cell functions could have been compromised under the 6H or ST conditions. Long-term culture conditions, perhaps favoring primitive hematopoietic cell cycling while preserving cell numbers and hopefully differentiation capacity, may offer an advantage over the short in vitro culture conditions used here.7 25
Characteristics of CD34-Enriched and Transduced Cells
| . | % CD34+ Post Column Selection . | % CD34+ Post Transduction . | CD34+ Cells Post:Pre Transduction . |
|---|---|---|---|
| Peripheral blood | |||
| 6H | 70.8 (14.5) | 65 (19.4) | 0.5 (14) |
| ST | 50 (29) | 38 (33) | 0.44 (10) |
| IL-3, 6, SCF | 53.2 (29) | 64.7 (26) | 1.1 |
| Bone Marrow | |||
| 6H | 67.6 (16) | 58.7 (26) | 0.43 (30) |
| ST | 67 (15) | 53 (8.5) | 0.62 (23) |
| IL-3, 6, SCF | 64 (11) | 73.3 (5.2) | 1.2 |
| . | % CD34+ Post Column Selection . | % CD34+ Post Transduction . | CD34+ Cells Post:Pre Transduction . |
|---|---|---|---|
| Peripheral blood | |||
| 6H | 70.8 (14.5) | 65 (19.4) | 0.5 (14) |
| ST | 50 (29) | 38 (33) | 0.44 (10) |
| IL-3, 6, SCF | 53.2 (29) | 64.7 (26) | 1.1 |
| Bone Marrow | |||
| 6H | 67.6 (16) | 58.7 (26) | 0.43 (30) |
| ST | 67 (15) | 53 (8.5) | 0.62 (23) |
| IL-3, 6, SCF | 64 (11) | 73.3 (5.2) | 1.2 |
Abbreviations: 6H, cells transduced for 6 hours without growth factors; ST, cells transduced for 72 hours in the presence of autologous stroma; IL-3, 6, SCF, cells transduced for 72 hours in the presence of IL-3, IL-6, and SCF.
Data from Dunbar et al.4
The inclusion of an autologous stromal layer during transduction was prompted by previous evidence that stromal cells or stromal matrix molecules can substitute for or enhance the effects of exogenous growth factors and improve gene transfer into primitive hematopoietic cells in animal models and human cells assayed in vitro.6,8,26 No toxicity was observed in manipulating patient stroma during transduction and on return of gene-marked cells and stroma to the patient. As the stroma was irradiated before use, is seems unlikely that any gene-marked stroma would contribute to endogenous stromal elements even if it reached the marrow in large numbers. The cytokines and matrix molecules elaborated by stromal cells would be predicted to have their greatest effects on the primitive hematopoietic cells during transduction. Such a growth environment is unlikely to enhance the growth of contaminating BC cells.27 Stromal may be more beneficial to the growth of contaminating myeloma.28 Previous attempts to perform gene transfer into myeloma cells have shown that myeloma cells are fragile and poorly tolerate in vitro handling and incubation.29 It is possible that the overall poor gene transfer efficiency to the long-term repopulating hematopoietic cells using stroma alone might be enhanced by the addition of growth factors during transduction.30
We would like to again stress the importance of marking studies as indices for new gene transfer vectors and transduction conditions. No in vitro assays have yet proven predictive of gene transfer efficiency to human long-term repopulating cells. Additionally, as our assumptions about gene transfer efficiency largely rest on in vitro studies of progenitor populations which may not reflect the dynamics of the most primitive cells, we may make false assumptions about the best course for future gene-marking studies. Better understanding and modeling of these most primitive interactions will be necessary before rational proposals for further marking trials are made.
ACKNOWLEDGMENT
The authors gratefully acknowledge the technical assistance of S. Sellers and members of the Cell Processing Laboratory in the Department of Transfusion Medicine. We also thank N.S. Young, A.J. Barrett, W. Wilson, A.W. Neinhuis, the members of the clinical care team, and our supportive research nurses: S. Phang, M. Caples, J. Kimball, N. McAtee, and M.A. Noone.
Address reprint requests to R.V.B. Emmons, MD, University of Massachusetts Medical Center, Division of Hematology/Oncology, 55 Lake Ave N, Room s7713, Worcester, MA 01655-0271.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal