Abstract
Neutrophil elastase (NE) is a serine protease that is transcriptionally regulated during early myeloid differentiation. The murine NE (mNE) promoter contains functionally important c-Myb, C/EBP, and ets binding sites. Deletion of the ets site reduced promoter activity by 90%. Although the ets transcription factor, PU.1, bound to this ets site, it only modestly activated the mNE promoter. Here, we show that a second transcription factor from myeloid cells — GABP — binds to the mNE ets site but strongly activates the mNE promoter. GABP is a heteromeric transcription factor complex that consists of GABPα, an ets factor, and GABPβ, a Notch-related protein. GABPα bound to the mNE ets site and, in turn, recruited GABPβ to form a transcriptionally active complex. GABPα and PU.1 competed with each other for binding to the mNE ets site. GABP increased the activity of the mNE promoter sevenfold in U937 myeloid cells. GABP cooperated with c-Myb and C/EBPα to activate the mNE promoter more than 85-fold in otherwise nonpermissive, nonhematopoietic NIH 3T3 cells. Thus, GABP binds to the crucial mNE promoter ets site and powerfully activates its expression alone and in cooperation with the transcription factors c-Myb and C/EBP.
GENE TRANSCRIPTION is an important regulatory mechanism in the differentiation of granulocytes and monocytes (myeloid cells) from immature bone marrow precursors. The crucial role of transcription in myeloid differentiation is indicated by the observation that many forms of myeloid leukemia, which represent disordered myeloid differentiation, are caused by aberrant expression of transcription factors. Myeloid genes are controlled by combinatorial patterns of lineage-restricted and more widely expressed transcription factors.1
The ets factors are DNA binding proteins that function as transcriptional activators that have been implicated in cellular growth control and differentiation. Myeloid cells express several ets factors, including PU.1,2-5 Ets-1,6,7 Ets-2,8 and GABP.9 Because ets transcription factors bind to related DNA sequences, it can be difficult to determine which specific ets factor is responsible for controlling the expression of particular myeloid genes. PU.1 binds to the promoters of CD11b,10 CD18,11 and other myeloid genes.12-18 Although PU.1 is essential for development of myeloid cells,19,20 its disruption did not fully abrogate the expression of genes thought to be regulated by PU.1.21 Thus, other ets factors may be responsible for transcriptionally activating genes that are considered targets of PU.1. We have shown that both PU.1 and GABP bind to crucial ets sites in the CD18 gene, but only GABP directly activated expression of CD18 in myeloid cells.9
Neutrophil elastase (NE) is a myeloid-restricted protease that is transcriptionally regulated early in myeloid differentiation.22 Because it is expressed in a lineage-specific and stage-specific pattern, NE is an excellent model for the molecular events that control early myeloid gene transcription. A 91-nucleotide (nt) fragment of the murine NE (mNE) promoter, which is sufficient to direct myeloid-specific gene expression, contains functionally important binding sites for c-Myb, C/EBP, and CBF. The mNE promoter also contains an ets site whose disruption reduced promoter activity by 90%.23 Although PU.1 from myeloid cells bound to this ets site, PU.1 activated mNE expression only modestly.24
Here, we demonstrate that the ets factor, GABPα, binds avidly to the mNE ets site. GABPα and PU.1 are the only ets factors from myeloid nuclear extracts that bind to the mNE promoter. GABPα recruits an unrelated protein, GABPβ, and together they form a heterodimeric complex on the mNE promoter. In contrast to PU.1, GABP strongly activates transcription of mNE in myeloid cells. Furthermore, GABP cooperates with c-Myb and C/EBP to powerfully activate mNE expression in otherwise nonpermissive, nonhematopoietic cells. Thus, mNE is the second myeloid gene, following our previous studies of CD18,9 in which the promoter is shown to be bound by both PU.1 and GABP, but whose myeloid transcription is activated only by GABP.
MATERIALS AND METHODS
Cell culture and transfection.U937 (ATCC #CRL 1593) cells were passaged twice weekly in RPMI 1640 (GIBCO BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (ICN, Costa Mesa, CA) in an atmosphere of 5% CO2 . NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium with 10% calf serum.
U937 cells were transfected as previously described25 with 20 μg mNE reporter constructs (pNE(Δ-91)LUC or pNE(Δ-79)LUC,24 5 μg of the empty expression vector pCAGGS26 or expression vectors for the human homologs of GABPα and GABPβ1-1 (pCAGGS-E4TF1-60 and pCAGGS-E4TF1-53, respectively),27 and 1 μg cytomegalovirus (CMV)/human growth hormone (hGH) (as an internal control for transfection efficiency). Promoter activity is expressed as normalized relative light units as previously described,11 and transfection results represent the mean ± SEM from at least three separate experiments, each in duplicate.
NIH 3T3 cells were transfected by calcium-phosphate precipitation, followed 18 hours later by a 3.5-minute, 15% glycerol shock. NIH 3T3 cells were transfected with 3 μg pNE(Δ-91)LUC or pNE(Δ-79)LUC and 0.25 μg pCAT-Control (Promega Corp, Madison, WI) as an internal control for transfection efficiency. Where indicated, cells were cotransfected with 0.1 μg each of expression vectors for GABPα and GABPβ1-1, already described, or pCAGGS empty vector; pMSV-C/EBPα (0.04 μg); and pCMV-Myb (0.5 μg).24 Transfections were performed in triplicate. Cells were harvested 36 to 42 hours later, and luciferase activity was normalized with pCAT-Control.
Preparation of nuclear extracts and proteins.U937 nuclear extracts were prepared as previously described.11 Glutathione S transferase (GST)-PU.1 (gift from Tony Kouzarides, Wellcome/CRC Institute, Cambridge, UK), GST-GABPα, and GST-GABPβ1-1 (E4TF1-60 and E4TF1-53, respectively; gifts from Hiroshi Handa, Tokyo Institute of Technology, Yokohama, Japan) were expressed in Escherichia coli, isolated with glutathione-sepharose, and quantified by staining with known protein standards in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), as previously described.9
Electrophoretic mobility shift assay.Figure 1 schematizes the mNE minimal promoter. The wild-type mNE probe (5′-GTGTCCCAGGGAGGAAGTAGGGCTG-3′ and its complement) corresponds to the -95/-71 region of the mNE promoter. The mutant mNE probe contains a GG → TT mutation in the underlined ets sequence. The oligonucleotides, which were synthesized with an additional TCGA at the 5′ end, were annealed and labeled with α32P-dCTP (ICN) by the Klenow fragment of DNA polymerase I (New England Biolabs, Beverly, MA). Radiolabeled probe (0.1 ng) was incubated for 10 minutes on ice with 1 μL purified protein or 10 μg nuclear extract in a 15-μL reaction containing 10 mmol/L Tris, pH 7.5, 50 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L β-mercaptoethanol, 1% Ficoll, and poly dI⋅dC (Pharmacia, Piscataway, NJ; 2.0 μg for nuclear extracts and 0.5 μg for purified proteins). Where indicated, one of the following unlabeled competitor DNA probes was added in a 100-fold molar excess: homologous mNE DNA, mutant mNE probe, or irrelevant probe (corresponding to -903/-883 of the CD18 promoter, which lacks an ets binding site).11 The ets factor competitive binding studies were performed with 1 μL GST-GABPα and the simultaneous addition of 1, 2, or 5 μL GST-PU.1; the converse experiment was performed with 1 μL GST-PU.1 and the simultaneous addition of 1, 2, or 5 μL GST-GABPα. Each fusion protein was estimated to be approximately 100 ng/μL based on Coomassie blue staining in SDS-PAGE. For supershift experiments, reactions were preincubated for 10 minutes on ice with 1 μL preimmune rabbit serum or 1 μL polyclonal rabbit antiserum raised against GABPα, GABPβ (both gifts from Tom Brown, Pfizer Inc, Groton, CT), or PU.1 (gift from Richard Maki, La Jolla Cancer Research Foundation, La Jolla, CA). Where indicated, 2 μg PU.1 competing peptide was included in the binding reaction. Products were electrophoresed at 180V in a 5% acrylamide/bis (19:1) gel in 45 mmol/L Tris base, 45 mmol/L boric acid, and 1 mmol/L EDTA (0.5× TBE) before autoradiography.
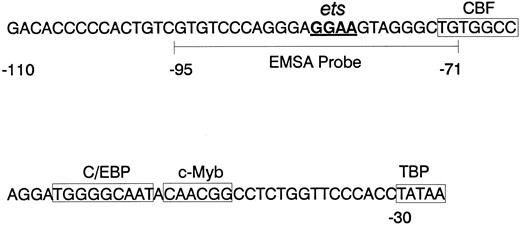
mNE promoter. The sequence of the proximal mNE promoter, which is sufficient to direct myeloid gene expression, is depicted. Boxes enclose binding sites for CBF, C/EBP, c-Myb, and TBP.23 24 The core GGAA sequence of the ets binding site is underlined, and the wild-type mNE ets probe (-95/-71) used for EMSA is bracketed.
mNE promoter. The sequence of the proximal mNE promoter, which is sufficient to direct myeloid gene expression, is depicted. Boxes enclose binding sites for CBF, C/EBP, c-Myb, and TBP.23 24 The core GGAA sequence of the ets binding site is underlined, and the wild-type mNE ets probe (-95/-71) used for EMSA is bracketed.
RESULTS
GABP binds to the mNE ets site.We prepared a double-stranded oligonucleotide probe that corresponds to -95/-71 of the mNE promoter (Fig 1) for use in the electrophoretic mobility shift assay (EMSA). This probe contains a GGAA motif that is a core binding sequence for numerous ets transcription factors, including PU.1 and GABP.
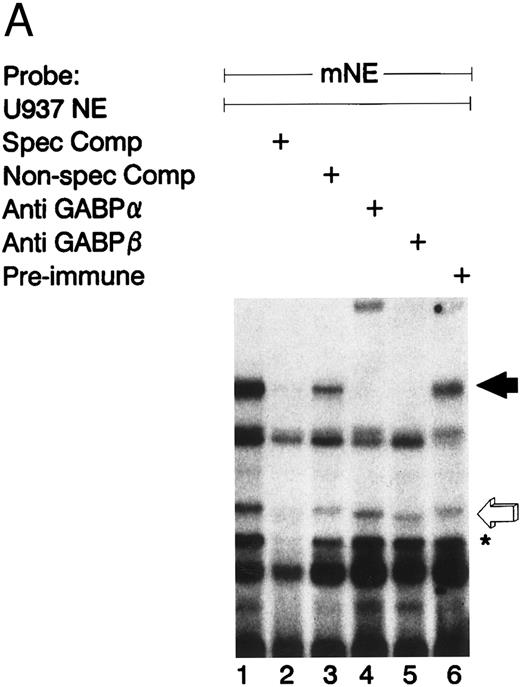
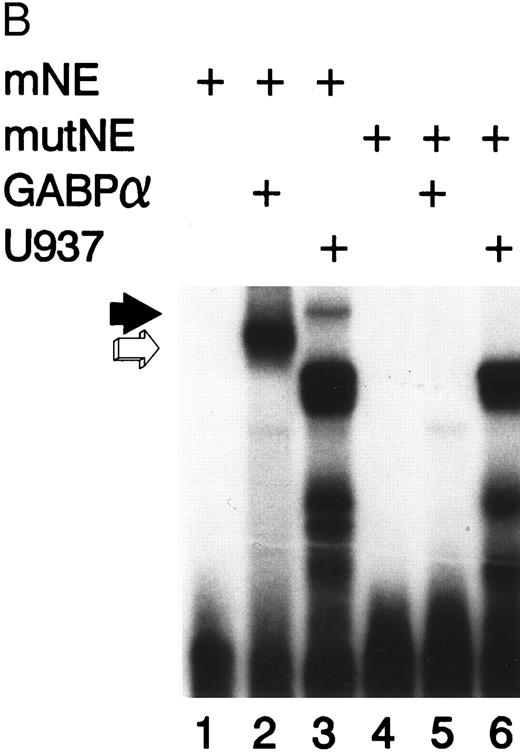
Figure 2A illustrates an EMSA with nuclear extract from U937 myeloid cells and the radiolabeled mNE -95/-71 probe. A species of low electrophoretic mobility bound to the mNE probe (lane 1, filled arrow). Binding by this species was sequence-specific, since it was abrogated by a 100-fold molar excess of unlabeled homologous probe (lane 2) but not by an irrelevant probe that lacks an ets binding site (lane 3). We performed an EMSA with specific antibodies to confirm the identify of the low-mobility binding complex as GABP. This binding species was unaffected by irrelevant antiserum (lane 6), but antibody to GABPα and GABPβ (lanes 4 and 5, respectively) abrogated this complex; antibody to GABPα generated a supershifted complex (lane 4). Thus, the low-mobility binding species contains both GABPα and GABPβ.
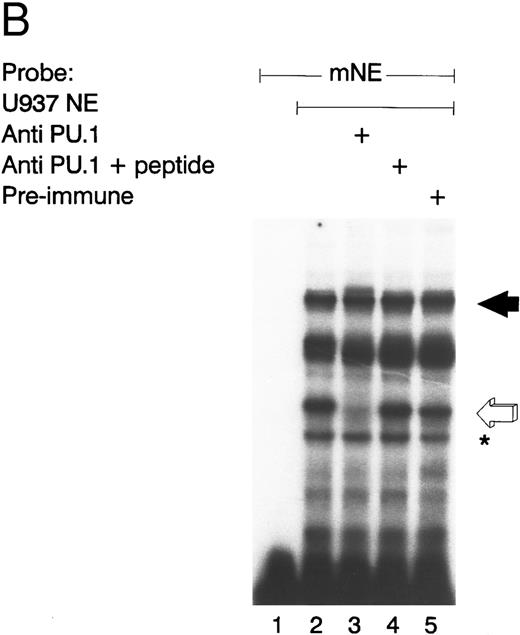
GABP from U937 myeloid nuclear extract binds to the mNE promoter. (A) A double-stranded oligonucleotide probe corresponding to -95/-71 of the mNE promoter was radiolabeled with 32P-dCTP. EMSA was performed with 10,000 cpm probe and 10 μg U937 myeloid nuclear extract. A species from U937 cells bound to this probe (lane 1, filled arrow) and was competed for by a 100-fold molar excess of homologous unlabeled probe (lane 2) but not by a 100-fold excess of irrelevant probe (lane 3). Binding of this species was abrogated by antibody to either GABPα or GABPβ (lanes 4 and 5, respectively) but not by preimmune serum (lane 6). Binding by PU.1 (open arrow) and a proteolytic degradation product of PU.1 (asterisk) was also seen. (B) EMSA was performed with the radiolabeled -95/-71 mNE probe and U937 nuclear extract. Antibody to PU.1 abrogated binding by PU.1 (lane 3), and this effect was eliminated by inclusion of PU.1 peptide in the binding reaction (lane 4); preimmune serum had no effect on its binding (lane 5).
GABP from U937 myeloid nuclear extract binds to the mNE promoter. (A) A double-stranded oligonucleotide probe corresponding to -95/-71 of the mNE promoter was radiolabeled with 32P-dCTP. EMSA was performed with 10,000 cpm probe and 10 μg U937 myeloid nuclear extract. A species from U937 cells bound to this probe (lane 1, filled arrow) and was competed for by a 100-fold molar excess of homologous unlabeled probe (lane 2) but not by a 100-fold excess of irrelevant probe (lane 3). Binding of this species was abrogated by antibody to either GABPα or GABPβ (lanes 4 and 5, respectively) but not by preimmune serum (lane 6). Binding by PU.1 (open arrow) and a proteolytic degradation product of PU.1 (asterisk) was also seen. (B) EMSA was performed with the radiolabeled -95/-71 mNE probe and U937 nuclear extract. Antibody to PU.1 abrogated binding by PU.1 (lane 3), and this effect was eliminated by inclusion of PU.1 peptide in the binding reaction (lane 4); preimmune serum had no effect on its binding (lane 5).
We detected PU.1 as a second probe-specific binding species from U937 cell nuclear extracts (Fig 2A, lane 1, open arrow). PU.1 from 32D cl3 murine myeloid cells had previously been shown to bind this probe.24 We confirmed the identity of this binding species from U937 cells by performing a supershift EMSA (Fig 2B). Antibody to PU.1 abrogated binding by this species (compare lane 3 v lane 2, open arrow), and this effect was reversed by inclusion of PU.1 peptide in the binding reaction (lane 4); no abrogation of PU.1 binding was seen with irrelevant antiserum (lane 5). A proteolytic degradation product of PU.1 is indicated by the asterisk, and it was unaffected by antibody to PU.1, as previously reported.11 No other sequence-specific binding species bound to this probe, indicating that GABP and PU.1 are the only ets factors from U937 myeloid cells that bound to the mNE ets site.
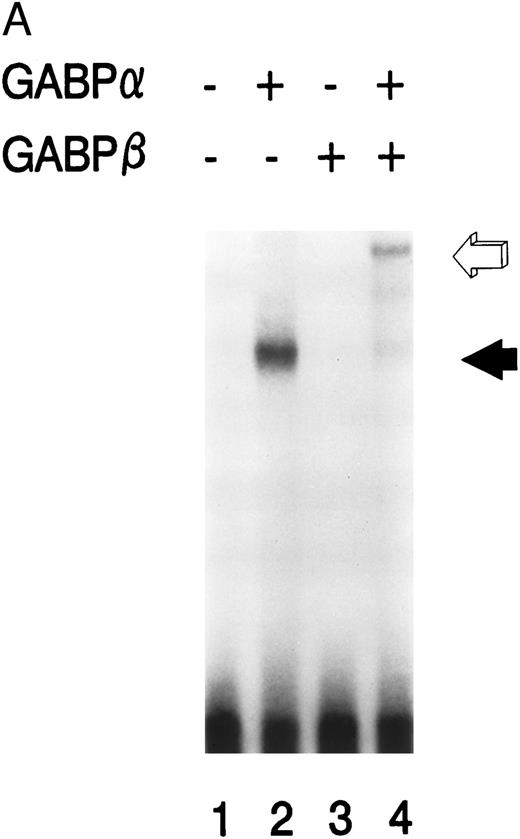
GABPα and GABPβ form a heterodimer on the mNE promoter.We prepared GABPα and GABPβ as GST fusion proteins for use in EMSA. Purified GABPα bound to the radiolabeled mNE -95/-71 promoter probe (Fig 3A, lane 2, filled arrow). No binding was seen with no added protein (lane 1) or with GABPβ alone (lane 3). Addition of both GABPα and GABPβ generated a complex of lower electrophoretic mobility (lane 4, open arrow) that corresponds to a GABPα/β heterodimer. GABP forms tetramers on promoters such as CD18 that contain more than one ets binding site,9 but we detected no GABP tetramers on the mNE promoter probe.
GABP forms a heterodimeric complex on the mNE promoter. (A) GABPα and GABPβ were prepared as GST fusion proteins. EMSA was performed with the mNE promoter probe and purified bacterially expressed GABPα (lanes 2 and 4) and GABPβ (lanes 3 and 4). Filled arrow, binding by monomeric GABPα; open arrow, a heterodimeric complex formed by GABPα and GABPβ. (B) A mutation of the ets site (GGAA → TTAA) was incorporated into the mNE promoter probe. EMSA was performed with the wild-type probe (mNE, lanes 1 to 3) or with the mutated probe (mutNE, lanes 4 to 6) and purified GST-GABPα (lanes 2 and 5) or U937 nuclear extract (lanes 3 and 6). Open arrow, binding by GST-GABPα that was disrupted by the ets site mutation (compare lane 2 and lane 5). Filled arrow, binding by GABP from U937 nuclear extract that was disrupted by the mutation (compare lane 3 and lane 6).
GABP forms a heterodimeric complex on the mNE promoter. (A) GABPα and GABPβ were prepared as GST fusion proteins. EMSA was performed with the mNE promoter probe and purified bacterially expressed GABPα (lanes 2 and 4) and GABPβ (lanes 3 and 4). Filled arrow, binding by monomeric GABPα; open arrow, a heterodimeric complex formed by GABPα and GABPβ. (B) A mutation of the ets site (GGAA → TTAA) was incorporated into the mNE promoter probe. EMSA was performed with the wild-type probe (mNE, lanes 1 to 3) or with the mutated probe (mutNE, lanes 4 to 6) and purified GST-GABPα (lanes 2 and 5) or U937 nuclear extract (lanes 3 and 6). Open arrow, binding by GST-GABPα that was disrupted by the ets site mutation (compare lane 2 and lane 5). Filled arrow, binding by GABP from U937 nuclear extract that was disrupted by the mutation (compare lane 3 and lane 6).
Mutation of the mNE ets site disrupts GABP binding.We incorporated the same GGAA → TTAA mutation into the mNE ets site that reduced myeloid activity of the mNE promoter by 90%,24 to determine if this mutation disrupts binding by GABP. Binding to the mNE probe by purified GABPα (Fig 3B, lane 2, open arrow) was abrogated by mutation in the ets site (lane 5). Similarly, binding to the wild-type probe by GABP from U937 nuclear extract (lane 3, filled arrow) was abrogated by the ets site mutation (lane 6). The intense doublet that is evident in lanes 3 and 6 (which was also observed in Fig 2) represents nonspecific binding to the probe and was unaffected by the ets site mutation. Thus, the same mutation that severely reduced myeloid activity of the mNE promoter abrogated binding by GABPα.
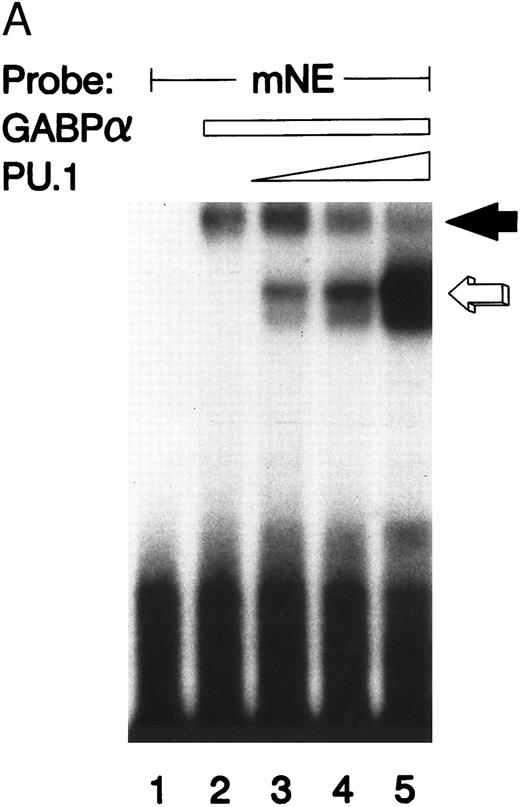
GABPα and PU.1 compete for the same mNE ets site.We performed an EMSA with GABPα and PU.1 prepared as GST fusion proteins. It should be noted that these binding species migrate more slowly than those formed by the corresponding native species from U937 nuclear extracts (compare with Fig 2) because the GST moiety contributes to the increased size of these fusion proteins. The mNE promoter probe was bound by both GST-GABPα (Fig 4A, solid arrow) and GST-PU.1 (open arrow). As increasing amounts of PU.1 were added, it competed with GABPα for binding to the mNE probe (lanes 3 through 5). No additional or novel complex was formed by addition of both GABPα and PU.1, indicating that under these conditions, no higher-order complex formed between these two ets factors. Thus, PU.1 and GABP bind to the same ets site in the mNE promoter.
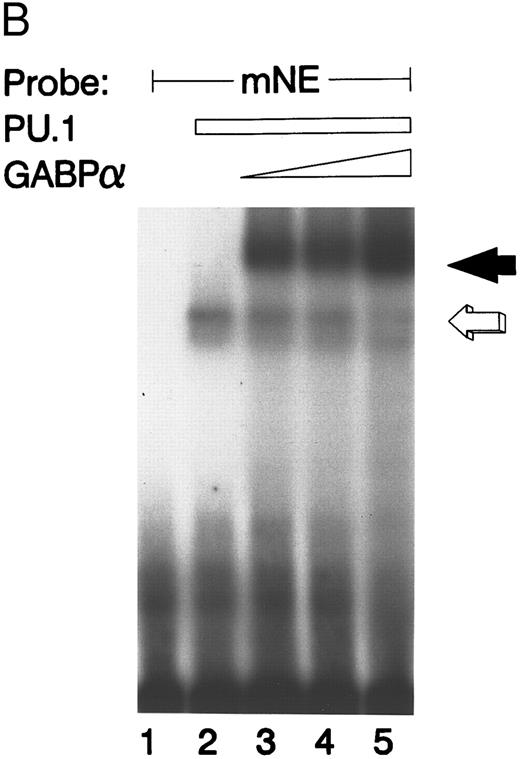
GABP and PU.1 compete for binding to the mNE promoter ets site. (A) EMSA was performed with the mNE promoter probe and purified bacterially expressed GST-GABPα and GST-PU.1. Filled arrow, binding by GABPα (lanes 2 to 5). Addition of increasing amounts of PU.1 generated a new binding complex (open arrow, lanes 3 to 5) that competed with GABPα for the radiolabeled mNE promoter probe. No novel complex that might represent a direct physical interaction of GABPα and PU.1 was seen. (B) Open arrow indicates binding by PU.1 (lanes 2 to 5). Addition of increasing amounts of GABPα (filled arrow, lanes 3 to 5) competed with PU.1 for the radiolabeled mNE promoter probe.
GABP and PU.1 compete for binding to the mNE promoter ets site. (A) EMSA was performed with the mNE promoter probe and purified bacterially expressed GST-GABPα and GST-PU.1. Filled arrow, binding by GABPα (lanes 2 to 5). Addition of increasing amounts of PU.1 generated a new binding complex (open arrow, lanes 3 to 5) that competed with GABPα for the radiolabeled mNE promoter probe. No novel complex that might represent a direct physical interaction of GABPα and PU.1 was seen. (B) Open arrow indicates binding by PU.1 (lanes 2 to 5). Addition of increasing amounts of GABPα (filled arrow, lanes 3 to 5) competed with PU.1 for the radiolabeled mNE promoter probe.
We performed the converse experiment with purified GST-PU.1 and GST-GABPα. PU.1 bound to the mNE probe (Fig 4B, open arrow), but the addition of increasing amounts of purified GABPα competed for binding to the mNE promoter probe (lanes 3 to 5, filled arrow).
GABP activates transcription of mNE promoter in myeloid cells.Because GABP from myeloid cells bound to the mNE promoter, we sought to determine if it functionally activates mNE expression. We transfected U937 myeloid cells with the mNE minimal promoter, pNE(Δ-91)LUC, and with expression vectors containing GABPα and GABPβ or the empty expression vector (pCAGGS). Each sample was transfected with the internal control CMV/hGH to control for transfection efficiency.
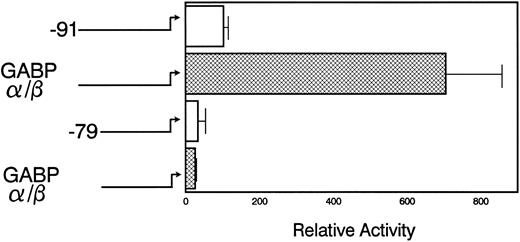
The pNE(Δ-91)LUC reporter was activated sevenfold when it was cotransfected with both components of the GABP complex (Fig 5). However, pNE(Δ-79)LUC, which lacks the mNE ets site, was not activated by GABP cotransfection. This indicates that GABP activation of mNE is dependent on the ets binding site. Cotransfection with either GABPα or GABPβ alone failed to activate the mNE promoter (data not shown). Although PU.1 from myeloid cells bound to the same ets site in the mNE promoter as GABP, cotransfection of pNE(Δ-91)LUC with PU.1 into myeloid cells activated the promoter less than twofold (data not shown). Thus, as we have found with the CD18 promoter,9 GABP is considerably more active than PU.1 for activating crucial ets sites to which both of these factors bind.
GABP activates myeloid transcription of mNE. U937 myeloid cells were transfected with 20 μg of either pNE(Δ-91)LUC (□) or pNE(Δ-79)LUC (▩), 5 μg each of pCAGGS-GABPα and pCAGGS-GABPβ or pCAGGS empty vector, and CMV/hGH (internal control). Luciferase activity was measured 14 hours after transfection and was normalized to GH expression. Data from 3 independent experiments in duplicate are shown as the mean ± SEM.
GABP activates myeloid transcription of mNE. U937 myeloid cells were transfected with 20 μg of either pNE(Δ-91)LUC (□) or pNE(Δ-79)LUC (▩), 5 μg each of pCAGGS-GABPα and pCAGGS-GABPβ or pCAGGS empty vector, and CMV/hGH (internal control). Luciferase activity was measured 14 hours after transfection and was normalized to GH expression. Data from 3 independent experiments in duplicate are shown as the mean ± SEM.
GABP cooperates with c-Myb and C/EBP to activate mNE in nonhematopoietic cells.PU.1 alone activated the mNE promoter only twofold in NIH 3T3 cells, either alone or cooperatively with c-Myb and C/EBP.24 Because GABP was considerably more effective than PU.1 for activating the mNE ets site in U937 myeloid cells, we reasoned that it might have a more powerful synergistic effect with c-Myb and C/EBP to activate mNE transcription.
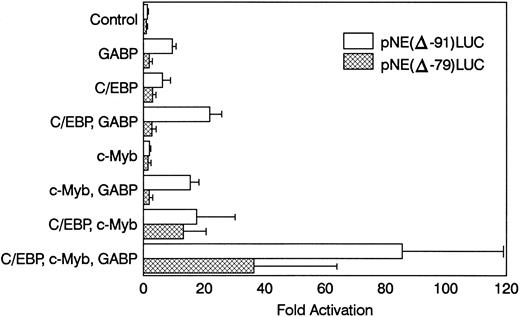
NIH 3T3 cells were cotransfected with pNE(Δ-91)LUC and expression vectors for GABPα/β, c-Myb, and C/EBPα (Fig 6). GABPα/β alone activated the mNE reporter 10-fold versus transfection with the empty pCAGGS expression vector. C/EBPα alone activated pNE(Δ-91)LUC sixfold. Cotransfection with GABPα/β and C/EBPα together resulted in 22-fold promoter activation. Transfection with c-Myb alone activated the mNE promoter only twofold, but cotransfection with GABP and c-Myb resulted in 15-fold activation. Transfection with C/EBPα and c-Myb activated pNE(Δ-91)LUC 18-fold. Transfection of pNE(Δ-91)LUC with GABPα/β, c-Myb, and C/EBPα resulted in greater than 85-fold promoter activation. Thus, each of these transcription factors individually activated mNE in NIH 3T3 cells. However, together they resulted in a multiplicative effect on mNE transcription and synergistically increased expression from this promoter.
GABP cooperates with c-Myb and C/EBPα to activate mNE transcription. NIH 3T3 cells were transfected with 3 μg pNE(Δ-91)LUC (□) or pNE(Δ-79)LUC (▩) and 0.25 μg pCAT-Control as an internal control for transfection efficiency. Where indicated, cells were cotransfected with pCAGGS-GABPα and pCAGGS-GABPβ (0.1 μg each) or pCAGGS empty vector, pMSV-C/EBPα (0.04 μg), and pCMV-Myb (0.5 μg). Data from 3 independent experiments are shown as the mean ± SEM.
GABP cooperates with c-Myb and C/EBPα to activate mNE transcription. NIH 3T3 cells were transfected with 3 μg pNE(Δ-91)LUC (□) or pNE(Δ-79)LUC (▩) and 0.25 μg pCAT-Control as an internal control for transfection efficiency. Where indicated, cells were cotransfected with pCAGGS-GABPα and pCAGGS-GABPβ (0.1 μg each) or pCAGGS empty vector, pMSV-C/EBPα (0.04 μg), and pCMV-Myb (0.5 μg). Data from 3 independent experiments are shown as the mean ± SEM.
When the same transfections were performed with the pNE(Δ-79)LUC reporter construct that lacks the crucial ets site, promoter activation was considerably blunted. Activation of pNE(Δ-79)LUC by C/EBPα and c-Myb individually (threefold and twofold, respectively) and together (13-fold) was relatively unaffected. However, GABPα/β alone failed to activate pNE(Δ-79)LUC (<twofold), and its cooperative effect with either C/EBP or c-Myb was considerably blunted (<threefold and <twofold, respectively). GABP modestly enhanced the effect of c-Myb and C/EBP on pNE(Δ-79)LUC despite the loss of the ets binding site. This activation may be due to a cryptic ets site in the construct, effects of GABP on the c-Myb or C/EBP expression vectors, or cooperative effects of GABP with these factors that do not require binding to DNA by GABP. Nevertheless, this effect was considerably reduced compared with activation of pNE(Δ-91)LUC by all three factors.
DISCUSSION
NE is a myeloid-specific gene that is transcriptionally regulated in early myeloid cells. We are studying mNE to define the DNA elements and transcription factors that control gene expression during early myeloid differentiation. As with other myeloid genes, expression of mNE is regulated by combinations of transcription factors that function synergistically, for their collective ability to activate mNE transcription is greater than the sum of their individual effects.28
A 91-nt fragment of the mNE promoter that is sufficient to direct myeloid-specific gene expression contains binding sites for c-Myb, C/EBP, and CBF. This minimal promoter fragment also contains an ets site whose disruption reduced promoter activity by 90%.23 We have now shown that GABPα from myeloid cells bound avidly to the crucial ets site in the mNE promoter. GABPα recruited the Notch-related protein, GABPβ, and together they formed a transcriptionally active heterodimeric complex on the mNE promoter. GABP and PU.1 are the only ets factors from myeloid nuclear extracts that bind to the mNE promoter. In contrast to PU.1, which only weakly activated the mNE promoter,24 GABP strongly activated mNE transcription in myeloid cells. Furthermore, GABP cooperated with c-Myb and C/EBP to increase mNE promoter activity greater than 85-fold in nonhematopoietic cells. Thus, although both GABP and PU.1 from myeloid cells bound to the mNE ets site, only GABP increased myeloid transcription of mNE.
GABP, which is also known as E4TF129 and NRF-2,30 consists of two distinct components. GABPα is an ets-related factor that binds to DNA, but alone it cannot activate gene transcription. It recruits its protein partner, GABPβ, and together they form a transcriptionally active multimeric protein complex. GABP forms tetrameric complexes (α2β2) on genes such as CD18 that contain two or more closely arrayed ets binding sites.9 We have previously shown that GABP activated myeloid transcription of CD18 greater than 10-fold, and together GABP and PU.1 cooperated to activate CD18 transcription in otherwise nonpermissive, nonhematopoietic cells.9 GABP also bound and activated the leukocyte-specific α4 integrin promoter31 and other viral and mammalian genes. Although GABP formed only a heterodimeric complex on the single ets site of the mNE promoter, this was sufficient for GABP to strongly activate its expression.
The mNE promoter represents the second myeloid gene to which both PU.1 and GABP from myeloid cells have been shown to bind. Their common sequence preferences suggest that other myeloid and lymphoid genes bound by PU.1 should be examined for binding and activation by GABP. However, competition by GABPα and PU.1 for binding to these sites in EMSA does not necessarily mean that they compete for binding in vivo. Their relatively equal affinities in EMSA for binding to mNE may not accurately reflect their properties in vivo, where cofactors may modulate their DNA binding affinities or affect their access to promoters. Alternatively, because these ets factors functionally cooperated to increase CD18 transcription in nonhematopoietic cells, they may play different roles in modulating gene expression. For example, one factor may alter chromatin configuration while the other actually activates transcription.
If the related ets factors PU.1 and GABP bind to a promoter, how can one determine which is responsible for activating gene transcription? PU.1 is an attractive candidate because it binds to multiple myeloid promoters,10-18 its expression is restricted to B-lymphoid cells and monocyte/macrophages,2 and disruption of the PU.1 gene in mice abrogates myelopoiesis.19,20 However, embryonic stem cells that are PU.1 null continued to express genes that are putative targets of PU.1. Notably, myeloperoxidase, which also contains regulatory ets sites and has early myeloid expression closely resembling that of mNE, was expressed by PU.1 null cells.21 The presence of GABP may account for the expression of myeloperoxidase and other early myeloid genes by PU.1 null cells.
The ets factors typically bind to DNA sequences containing a GGAA core, but they exhibit distinct preferences for flanking nts. The GGAGGAAGTA sequence of the mNE ets site closely resembles idealized GABP binding sites determined by mutagenesis studies (GGAAGTG)29 and binding site selection (GCCGGAAGT).32 An optimal PU.1 binding site is harder to define, for although most PU.1 binding sites share the sequence GAGGAA, some functional PU.1 elements lack even the supposedly canonical GGAA sequence.10 Although myeloid cells express other ets factors, GABP and PU.1 were the only ets factors we detected that bind to the mNE promoter ets site. For example, Ets-2 is present in myeloid cells,8 and its constitutive expression in a myeloblast cell line forces macrophage-like differentiation.33 However, Ets-2 bound only weakly to the mNE promoter; mutation of the mNE ets site to a more optimal Ets-2 consensus site (CCGGAACTA) enhanced its binding (data not shown). Furthermore, Ets-2 failed to activate mNE reporter genes in myeloid cells (data not shown), presumably because of its lower affinity for the mNE ets site. Thus, the ability of PU.1 and GABP to bind mNE is not a general property of ets factors, but instead may be determined by their distinct sequence preferences.
Although Ets-2 bound weakly to the mNE ets site, it did activate the mNE promoter individually (fivefold) and cooperatively with c-Myb and C/EBP (30-fold) following transfection into NIH 3T3 cells (I.N. and A.D.F., data not shown, March 1995). What might account for the discordance between the ability of Ets-2 to activate mNE in myeloid cells and nonhematopoietic cells? One possibility is that other cooperative factors might alter the cellular milieu. Recently, MafB was described as a lineage-restricted factor that inhibits Ets-1 transcriptional activation in myeloid cells but not in erythroid cells.7 One may speculate that MafB or other inhibitory factors could inhibit Ets-2 activation of mNE in myeloid cells, but its absence from nonhematopoietic cells could have permitted unopposed activation in NIH 3T3 cells.
Combinations of the factors that control mNE expression — c-Myb, C/EBP, and ets factors — have been implicated in controlling other myeloid genes. The myeloid-specific avian gene, mim-1, is controlled by c-Myb, an avian C/EBP equivalent (NF-M), and an ets factor. The granulocyte-macrophage colony-stimulating factor (GM-CSF ) receptor promoter is activated by C/EBPα and the ets factor, PU.117; c-Myb and an ets factor control CD13 expression34; and the M-CSF receptor promoter is activated by C/EBP, PU.1, and the CBF complex.35,36 The functional cooperation of Myb and ets factors is further suggested by the observation that a fusion of Myb and ets acts as an oncogene for avian leukemias, but the absence of either component renders this oncogene nontransforming.37 GABP and C/EBP were recently shown to cooperate in activating transcription from the promoter of the coagulation protein, factor IX.38
None of the factors that control mNE expression are restricted to myeloid cells. Myb is expressed by immature cells of all hematopoietic lineages.39 C/EBPα, the isoform most active for increasing mNE transcription, is most highly expressed in early myeloid cells.40 Although other C/EBP isoforms can activate mNE,24 their expression pattern contrasts with mNE, since their expression increases during myeloid differentiation.40 C/EBP is also expressed in nonhematopoietic cells, including hepatocytes41 and adipocytes.42 GABPα and GABPβ are expressed by both hematopoietic and nonhematopoietic cells.43 PU.1 expression is limited to B lymphocytes and myeloid cells,2 but its expression increases as myeloid cells differentiate,4 a pattern that differs distinctly from that of NE. Thus, even combinatorial patterns of these factors fail to fully explain the lineage restriction and the characteristic early myeloid transcription of NE.
Although the two GABP subunits are widely expressed by hematopoietic and other cell lineages, subtle alterations in GABP expression may contribute to lineage-restricted gene expression. GABPβ, the subunit that contributes the transcriptional activation and multimerization domains to the complex, may be differentially regulated. At least four alternately spliced isoforms of GABPβ are expressed,30 and an independent gene that is highly homologous to GABPβ has been cloned.44 Myeloid cells express at least four distinct GABPβ species, and these isoforms differentially activate the CD18 promoter (M.L. and A.G.R., unpublished observation, November 1995). Furthermore, specific GABPβ isoforms may be differentially expressed during myeloid differentiation (A.G.R., unpublished observation, January 1997). Thus, subtle modulations of GABP may contribute significantly to myeloid gene expression.
Posttranslational modifications of the factors that control mNE transcription may also contribute to its regulated myeloid expression. Phosphorylation modulates DNA binding and transcriptional activation by many transcription factors, including c-Myb,45 C/EBP,46 and the ets factor, Elk-1.47 Phosphorylation of PU.1 is crucial for its interaction with a B-lymphoid protein partner, NF-EM5/Pip,48,49 and there is recent evidence that PU.1 is phosphorylated in myeloid cells.50 GABP itself now appears to be a target for regulation via phosphorylation. GABP binds and activates the prolactin promoter, and insulin treatment of pituitary cells increases prolactin transcription and induces GABPα phosphorylation.51 Expression from the human immunodeficiency virus type 1 long terminal repeat (LTR) in Jurkat T cells is increased by the signal transduction protein, Raf-1, and this effect requires GABP binding. Overexpression of Raf-1 in Jurkat cells is associated with phosphorylation of both GABPα and GABPβ.52 We recently found that both GABPα and GABPβ are targets for phosphorylation by casein kinase II (data not shown). Thus, recent evidence demonstrates that GABP is phosphorylated in response to known signal transduction pathways. Future investigations may link signal transduction pathways with ets-regulated gene expression during myeloid differentiation.
ACKNOWLEDGMENT
The authors thank Hiroshi Handa for GABPα (E4TF1-60) and GABPβ (E4TF1-53) GST fusion and expression vectors, Tony Kouzarides for GST-PU.1, Tom Brown for antisera to GABPα and GABPβ, and Richard Maki for antiserum to PU.1.
Supported in part by an American Heart Association Rhode Island Affiliate Fellowship (C.P.S.), National Institutes of Health grant no. R01HL51388 (A.D.F.), and National Institute of Diabetes and Digestive and Kidney Diseases grant no. R29DK44728 (A.G.R.).
Address reprint requests to Alan G. Rosmarin, MD, The Miriam Hospital, Research 215, 164 Summit Ave, Providence, RI 02906.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal