Abstract
The recent finding of somatically mutated μ heavy chain transcripts in human peripheral blood (PB) B lymphocytes suggests that T-dependent B-cell memory might not be restricted to class-switched cells. We provide here evidence that IgM-only PB B cells are likely to be the IgM-expressing counterpart of classical (IgM−IgD−) memory B cells in humans. As shown by molecular single cell analysis, most IgM-only cells carry mutated V region genes, like class-switched cells. Although both subsets represent populations of nonactivated, resting cells, they express higher levels of Ig mRNA than naive (IgM+IgD+) B cells. IgM-only and class-switched cells are CD38−CD77−, and mostly CD23−, thus neither resembling germinal center nor naive B cells. Because many IgM-expressing B cells located in secondary lymphoid tissues resemble IgM-only PB B cells in terms of cell phenotype, we propose that the human lymphoid system contains a large compartment of IgM-expressing memory cells. Moreover, these cells seem to represent the nonmalignant counterparts of IgM-expressing tumor cells in sporadic Burkitt's lymphoma, MALT lymphoma, monocytoid B-cell lymphoma, and diffuse large-cell lymphoma that were found to harbor somatically mutated V genes.
BONE MARROW-DERIVED IgM+IgD+ cells are precursors of germinal center (GC) B cells that hypermutate their V region genes in the course of a T-cell–dependent immune response to foreign antigens.1-4 Studies in the mouse have shown that selected antibody mutants5,6 are released into the periphery as long-lived, isotype-switched memory B cells.7-9 Accordingly, the memory B-cell pool, responsible for an accelerated immune response upon renewed antigen encounter, would consist of somatically mutated, IgG-, IgE-, or IgA-expressing B cells. In the literature, one finds surprisingly little indication for an analogous IgM memory compartment.10-12 At least from a mechanistic point of view, there is no reason that memory B cells should exclusively express Ig heavy chains other than those of the μ class. It is known that somatic hypermutation and isotype switch are independent events,13 indicating that class-switch is neither a prerequisite for nor a consequence of affinity maturation. The recent findings of somatically mutated μ transcripts in the human peripheral blood (PB)14,15 provided a first indication that T-cell–dependent B-cell memory might not be restricted to class-switched cells. We subsequently showed that mutated μ transcripts in PB B cells are confined to the fraction of cells that express IgM but no or little IgD (IgM-only cells), whereas IgM+IgD+ B cells harbor unmutated V region genes.16
In the present work, we investigate whether these IgM-only PB B cells or a fraction of them resemble classical (IgM−IgD−) memory B cells7-9 in terms of the percentage of mutated cells within the subset, cell phenotype, and cell cycle distribution. We first determined the fraction of somatically mutated among all IgM-only cells by amplifying and sequencing VκJκ genes from single, flow cytometrically isolated cells. This excluded possible biases of population analyses in cDNA-based approaches caused by differing mRNA levels in B-cell subsets (see below). We then analyzed cell surface markers and the cell cycle distribution of the IgM-only cells and compared these characteristics with those of classical (IgM−IgD−) memory cells on the one hand and naive (IgM+IgD+) B cells as well as B cells proliferating in GCs on the other. Finally, we investigated whether IgM-only PB B cells contain elevated Ig mRNA levels compared with IgM+IgD+ B cells to explain the discrepancy between the large fraction of somatically mutated μ transcripts (up to 60%) in cDNA libraries generated from unseparated PB B lymphocytes on the one hand14,15,17,18 and the low percentage (12%) of IgM-only cells among IgM-expressing PB B lymphocytes on the other.16,19 20 For this purpose, we compared κ mRNA levels in IgM+IgD+, IgM-only, and, in addition, class-switched (IgG+ and IgA+) PB B cells.
MATERIALS AND METHODS
Cell separation and flow cytometry.Five hundred milliliters of heparinized PB from three normal, healthy donors was obtained from the blood bank of the Institut für Transfusionmedizin of the Cologne University Hospital (Cologne, Germany). PB mononuclear cells (PBMC) were isolated by Ficoll-Isopaque density centrifugation and CD19+ B cells were enriched to 95% to 98% by magnetic cell separation using the MiniMACS system (Miltenyi Biotec, Bergisch Gladbach, Germany) as described.16 The B-cell–enriched cell suspension was incubated with biotinylated goat antihuman (GaH)-IgD (Southern Biotechnology Associates [SBA], Birmingham, AL), GaH-IgM-phycoerythrin (PE) and GaH-IgA-fluorescein isothiocyanate (FITC) (Sigma, München, Germany), and GaH-IgG-FITC (SBA) for 15 minutes on ice. After washing with phosphate-buffered saline/1% bovine serum albumin/0.02% NaN3 , GaH-IgD was developed with Streptavidin-CyChrome (Pharmingen, San Diego, CA) for 10 minutes on ice. Cells were separated into IgM+IgD+, IgM++IgD−, and IgG+ and IgA+ fractions, and in one experiment also into an IgM++IgD+ fraction, on a FACS 440 (Becton Dickinson & Co, Mountain View, CA). Forward and side scatter parameters were set to exclude nonlymphocytes and cell debries. Dead cells were excluded by propidium iodide staining. In all sortings, 105 cells of each B-cell subset were collected in 1.5-mL Eppendorf reaction tubes. At least two tubes were prepared per subset. The purity of the sorted B-cell fractions was determined on a FACS 440.
For the detection of possibly contaminating plasma cells, 3 × 104 cells per B-cell subset were spun down on glass slides. Cells were fixed and incubated with anti-κ-FITC/anti-λ-PE (Becton Dickinson).
For the isolation of single κ-expressing IgM++IgD− cells, 20 mL heparinized PB was obtained from a healthy, 32-year-old donor. CD19+ B cells were enriched as described above. The B-cell–enriched cell suspension was incubated with GaH-IgD and GaH-IgM, as described above, and with FITC-conjugated GaH-Igk (Becton Dickinson). A fraction of PBMC was stained with anti-CD3-FITC (Becton Dickinson). On a FACS 440, single κ+ IgM++IgD− B cells and, in addition, single T cells (CD3+) were sorted directly into polymerase chain reaction (PCR) reaction tubes containing 20 μL PCR buffer and 0.05 μg 5s rRNA.
For the analysis of surface antigens, 5 × 105 cells of the CD19+-enriched cell suspension (95% to 99% purity) were incubated with GaH-IgD and GaH-IgM, as described above, and with either FITC-conjugated anti-CD25, anti-CD71, anti-CD5, anti-CD23, anti-CD10 (Becton Dickinson), or anti-CD38 or with unconjugated anti-CD77 (both Immunotech, Marseille, France), developed in a second staining step by mouse antirat IgM (Serotec, Oxford, UK). Ten thousand and, in the case of the anti-CD5 and anti-CD23 stainings, 5 × 104 events per staining were collected on a FACScan (Becton Dickinson).
For the cell cycle analysis, 40 mL PB was obtained from a healthy volunteer, and CD19+ B cells were isolated as described above. The CD19+-enriched fraction was incubated with anti-IgM-PE and anti-IgD-FITC (SBA). On a FACS 440, IgM++IgD− and IgM+IgD+ cells were sorted as described above. Isolated cells were subsequently lyzed in a 0.1% sodium citrate solution containing 0.05 mg/mL propidium iodide for 10 minutes.21 Cells of an Epstein-Barr virus (EBV) cell line were treated accordingly. A total of 2 × 104 events per sample were collected on a FACS 440.
RNA isolation and cDNA synthesis.Cellular RNA was isolated from 105 cells each of the IgM+IgD+, IgM++IgD−, IgG+ and IgA+, and IgM++IgD+ fractions, respectively, as described.22 RNA was prepared from the respective B-cell subsets of three donors (donors A, B and C). Total cellular RNA was hybridized to two oligonucleotides, one specifically hybridizing to the first exon of the κ chains constant region gene16 and the other to a sequence within the β-actin gene (3′β-actin: 5′-GGGAGGTAGCAGGTGGCGTTTACGAAGATC-3′). First-strand cDNA synthesis was performed using SuperScript Moloney's murine leukemia virus (MMLV) reverse transcriptase (GIBCO BRL, Grand Island, NY). Two cDNA mixtures of each B-cell subset were prepared from the IgM+IgD+, IgM++IgD−, and IgG+ and IgA+ fractions.
Cloning and sequencing of material amplified by PCR.One-twentieth of the first-strand cDNA mixtures of the IgM+IgD+, IgM++IgD−, IgG+ and IgA+, and IgM++IgD+ fractions from a 27-year-old donor (donor B) was amplified in separate PCR reactions using the Cκ primer and a 5′ Vκ3-framework region (FR) I primer.16 Amplification was performed in 50 μL reaction mixtures containing 10 mmol/L Tris-HCl, 50 mmol/L KCl, 2.5 mmol/L MgCl2 , 200 μmol/L dNTP, 0.125 μmol/L of each primer, and 1.7 U Taq polymerase. The amplification program consisted of 35 cycles of 60 seconds at 95°C, 30 seconds at 63°C, and 60 seconds at 72°C, followed by 5 minutes at 72°C. Taq polymerase was added after the first heating to 95°C. Amplifications were performed in a Trio Thermoblock (Biometra, Hannover, Germany). Gel-purification of PCR products and cloning was performed as described previously.16 Sequences were determined using the GATC 1500 direct blotting electrophoresis DNA sequencing system (MWG Biotech, Ebersberg, Germany). Inserts were sequenced using digoxigenin-labled primers and the DIG Taq DNA sequencing kit (Boehringer Mannheim, Mannheim, Germany). The Boehringer Nucleic Acid Detection Kit was used for colorimetric detection. Sequences were analyzed with DNASIS software (Pharmacia, Freiburg, Germany).
Single-cell PCR.Single cells in PCR buffer were incubated with 0.25 mg/mL proteinase K for 50 minutes at 50°C. Inactivation of the enzyme was performed by incubating the mixture at 95°C for 8 minutes. For the first round of amplification, a 5′Vκ-primer mix consisting of four oligonucleotides [Vκ1, 2, 423; Vκ3: 5′-TTGTG(A/T)TGAC(A/G)C-AGTCTCCAG(G/C)CACC-3′] that hybridize specifically to the corresponding sequences in the FRI of all members of the Vκ1-4 gene families and a 3′Jκ5 primer [5′-AAATGCTT(A/T)CGTTTAATCTCCAGTCG-3′] were used. The first round was performed in the same reaction tube in a 50 μL volume using the conditions described in the previous section, except that the MgCl2 concentration was 1.5 mmol/L. For the second round of PCR, the Vκ family-specific primers and a nested Jκ primer mix consisting of Jκ family-specific primers (Jκ1, 2, 4; Jκ3; and Jκ5 [outer Jκ primers4]) were applied. The second round of amplification was performed in separate reactions for each of the four Vκ family-specific primers using 0.5 to 1.0 μL of the first-round reaction mixture in a 50 μL volume using the conditions described above (1.5 mmol/L MgCl2 ). A 10-μL aliquot was analyzed on a 2% agarose gel. One microliter of the reaction mixtures giving rise to a PCR product was amplified in separate reactions with the respective Vκ family-specific primer and with each of the Jκ family-specific primers (outer or inner Jκ-primers4 ). PCR products were gel-purified. An aliquot of the isolated DNA was sequenced as described in the previous section, except that Vκ and Jκ family-specific digoxigenin-labeled primers were used. Because PCR products obtained from individual rearrangements can be directly sequenced without being cloned, sequence errors due to misincorporation by Taq polymerase are negligible.
Comparative PCR.One-twentieth of each first-strand cDNA mixture, corresponding to about 5,000 cell equivalents (CE), was amplified using the Cκ-primer and the 5′Vκ-primer mix. In separate PCR reactions, one-twentieth of a cDNA mixture was amplified using the 3′β-actin primer and a 5′β-actin primer (5′β-actin: 5′-ATCTACCCGTGTCACA-CCCACTGGGGCAGT-3′). From each cDNA mixture, two amplifications per primer combination (Vκ-mix/Cκ and 5′/3′β-actin) were performed. Except for the number of cycles and the MgCl2 concentration (1.5 mmol/L), the conditions for amplification were as described above. Twenty and 23 cycles of amplification were performed for the corresponding samples from donor A and donors B and C, respectively. Water controls were included in each experiment.
One-fifth of each PCR mixture was loaded onto a 2% agarose gel. After gel-electrophoresis, PCR products were transfered to a Nytran filter according to standard procedures.24 Filters were hybridized with 32P-labeled PCR-products. For the generation of radioactively labeled probes, 4 μL of purified PCR product (1/100 dilution) obtained by amplifying PB lymphocyte cDNA with the Vκ-mix/Cκ or the 5′/3′β-actin primer combination, respectively, was amplified for 30 cycles under the conditions described in the previous section. A total of 40 μCi α32P dATP (3,000 Ci/mmol) was added to each PCR reaction mixture. The labled PCR-products were isolated according to standard procedures.24 After hybridization with the labeled PCR products, final washing of the corresponding filters was performed under the following conditions: 0.1× SSC/0.1% sodium dodecyl sulfate at 52°C. Densitometric analysis of filters for quantification of band intensities was performed using a phosphoimager (Fuji, Tokyo, Japan).
Validity of the comparative PCR.To compare κ mRNA levels between separate PB B-cell subsets, we sorted equal cell numbers per B-cell subset (IgM+IgD+, IgM++IgD−, and IgG+ and IgA+) directly into reaction tubes followed by RNA isolation, cDNA synthesis, and PCR-amplification of κ transcripts. RNA isolation as well as cDNA synthesis of the fractions were performed in parallel to minimize variation between samples as a result of differing reaction conditions. The corresponding VκCκ as well as β-actin PCRs were also performed in parallel. The preparation of two separate cDNAs per B-cell subset and, in addition, of two separate PCR reactions per cDNA mixture should allow us to estimate the influence of sampling errors. In preliminary experiments, it had been tested out that the amplification is still in its exponential phase between 20 and 25 cycles (data not shown).
For the comparison of κ mRNA levels between PB B-cell subsets, we refered to a defined number of cells per subset instead of quantifying the VκCκ-PCR products by relating them to the corresponding cDNA amplifications of a constitutively expressed gene. It is known that the expression of houskeeping genes can vary with cell cycle or upon treatment with drugs.25 26 Therefore, it is possible that mRNA levels of constitutively expressed genes vary also between subpopulations of cells. Amplifications of β-actin transcripts were included in the analysis to test the integrity of the cDNA mixtures.
The Vκ1-Vκ4 FRI primers recognize nearly all Vκ germline genes.27 Therefore, differences in band intensities of VκCκ-PCR-products are unlikely to be due to differing Vκ gene usage between B-cell subpopulations.
We determined whether the finding of differences in terms of Ig mRNA levels between B-cell subsets can be reproduced using different amounts of starting material. Therefore, two samples each of IgD+ and IgD− PB B cells were sorted and RNA isolation and cDNA synthesis were performed as described above. From each of the mixtures, 20,000, 5,000, 2,500, 1,250, and 625 CE were amplified in separate PCRs (Vκ and β-actin); two reactions were performed for each cDNA mixture of the 20,000, 2500, and 625 CE samples, four for those of the 5,000 and 1,250 CE samples. Twenty thousand, 5,000, and 2,500 CE were amplified for 23, 1,250, and 625 CE for 25 cycles. After gel electrophoresis, PCR products were blotted, hybridized to radioactively labeled probes, and finally densitometrically analyzed as described above. κ mRNA quotients of IgD− (memory)/IgD+ cells of the respective samples were determined (β-actin quotients in brackets): 20,000 CE, 2.4 (1.3); 5,000 CE, 3.5 (0.9); 2,500 CE, 4.9 (0.9); 1,250 CE, 4.1 (1.0); and 625 CE, 4.5 (1.1). The lowest quotient of 2.4 determined for the 20,000 CE samples suggests that the PCR was already in a saturating phase, because the amount of starting material was too high. Although there is variation between the values of the κ mRNA quotients determined for 5,000, 2,500, 1,250, and 625 CE, they remain in a similar range over an eightfold range.
Overall, the experimental approach chosen in the present work should allow a comparison between κ mRNA levels in the three PB B-cell subsets analyzed.
Culturing of PB B-cell subsets.IgM-only, IgM+IgD+, and class-switched B cells were isolated as described above. Duplicates and, in some cases, triplicates of 105 cells of each subset were cultured in medium with a cocktail of interleukin-2 (IL-2), IL-10, and staphylococcus aureus concanavalin for 10 days in the CD40-system.28 Levels of IgG anti-tetanus toxoid (TT) antibodies and total IgG in the culture supernatants were quantified by enzyme-linked immunosorbent assay.28 Only in some supernatants (4/21) was the level of TT-specific antibodies found to be above background, despite normal total IgG levels. Also, there was no correlation between cultures giving rise to positive TT signals and a particular B-cell subset.
RESULTS
Somatically mutated V region genes in IgM-bearing PB B cells are confined to IgM++IgD− cells.Naive B cells (IgM+IgD+), IgM-only (IgM++IgD−), and class-switched (IgG+ and IgA+) B cells from a 27-year-old donor were purified after enrichment of CD19+ B cells by magnetic cell separation. In addition, B cells, which — like IgM-only cells — express high levels of surface IgM but also normal levels of IgD (IgM++IgD+), were isolated. Such cells had not been analyzed in our earlier work.16 Figure 1 shows the gates set for sorting and the reanalyses of sorted cells. Amplified Vκ3 cDNA from the four fractions was cloned into plasmids and nucleotide sequences of 27 inserts (4 IgM+IgD+, 4 IgG+ and IgA+, 8 IgM++IgD−, and 11 IgM++IgD+) were determined. Nine sequences could be assigned to A27, 9 to L6, 8 to L2, and one to L20 (for references see Schäble and Zachau27; data not shown). All sequences showed unique Vκ-Jκ junctions (data not shown). In agreement with our earlier data,16 18 inserts (67%) carried non–germline-encoded nucleotides at the Vκ-Jκ border. All but two of the Vκ rearrangements were in-frame.
Fluorescence analysis of B cells derived from PB lymphocytes of a 27-year-old person (donor B; left and middle) and a 32-year-old donor (right). (Top left) Anti-IgD/anti-IgM and (bottom left) anti-IgG and IgA/anti-IgM three color stainings of CD19+ B cells enriched by MACS and the corresponding reanalyses of (top middle left) IgM++IgD−, (top middle right) IgM+IgD+, (bottom middle left) IgG+ and IgA+, and (bottom middle right) IgM++IgD+ B-cell populations. Indicated are the gates set for sorting of the respective B-cell subsets. The sorted fractions derived from this donor (B) were used for the sequence analysis of expressed Vκ3 genes and, except the IgM++IgD+ fraction, for the comparative PCR experiment. (top right) Anti-IgD/anti-IgM and (bottom right) anti-κ stainings of CD19+ B cells enriched by MACS. Indicated are the gates set for sorting of single κ+ IgM++IgD− cells.
Fluorescence analysis of B cells derived from PB lymphocytes of a 27-year-old person (donor B; left and middle) and a 32-year-old donor (right). (Top left) Anti-IgD/anti-IgM and (bottom left) anti-IgG and IgA/anti-IgM three color stainings of CD19+ B cells enriched by MACS and the corresponding reanalyses of (top middle left) IgM++IgD−, (top middle right) IgM+IgD+, (bottom middle left) IgG+ and IgA+, and (bottom middle right) IgM++IgD+ B-cell populations. Indicated are the gates set for sorting of the respective B-cell subsets. The sorted fractions derived from this donor (B) were used for the sequence analysis of expressed Vκ3 genes and, except the IgM++IgD+ fraction, for the comparative PCR experiment. (top right) Anti-IgD/anti-IgM and (bottom right) anti-κ stainings of CD19+ B cells enriched by MACS. Indicated are the gates set for sorting of single κ+ IgM++IgD− cells.
For the determination of somatic mutation frequencies, nucleotide differences in the Vκ3 sequences relative to the corresponding Vκ3 and Jκ germline genes were considered. The fact that all human Vκ germline genes are known and that the individual germline genes seem to exhibit no or little polymorphism within the human population27 allows unambiguous identification of somatic mutations in rearranged Vκ genes. Table 1 summarizes the range of mutations per V region gene and the somatic mutation frequencies of the subsets analyzed. Both the number of mutations per mutated V region gene and the mutation frequencies determined for the IgM+IgD+ (0.2%), IgM-only (1.7%), and class-switched (4.2%) subsets were similar to those observed in the respective populations in our previous work (0.3%, 2.0%, and 3.9%, respectively16 ). Among 11 Vκ3 sequences derived from the IgM++IgD+ fraction, six were identical to and five showed between 1-and 6-bp differences (1, 1, 2, 4, and 6) to the corresponding Vκ3 and Jκ germline genes. The mutation frequency of this fraction is 0.4%. This value is considerably below that determined for the IgM-only subset (1.7%) and only slightly higher than the value determined for the naive IgM+IgD+ subset (0.2%). Thus, most of the PB B cells that express IgD as well as high levels of surface IgM (IgM++IgD+) appear to belong to the subset of naive B cells rather than to the fraction of IgM-only cells with which they share high surface IgM expression. The κ transcripts that show 4 and 6 mutations, respectively, might be derived from contaminating somatically mutated IgM-only cells.
Summary of Mutations in the V Regions Expressed in Vk-Bearing IgM+IgD+, IgM++IgD+, IgM-Only, and Class-Switched B Cells Derived From the PB of a 27-Year-Old Person
| Cell Population . | n* . | Range† . | % Mutation‡ . |
|---|---|---|---|
| IgM+IgD+ | 4 | 0-1 | 0.2 |
| IgM++IgD+ | 11 | 0-6 | 0.4 |
| IgM++IgD− | 8 | 0-12 | 1.7 |
| IgG+ and IgA+ | 4 | 3-23 | 4.2 |
| Cell Population . | n* . | Range† . | % Mutation‡ . |
|---|---|---|---|
| IgM+IgD+ | 4 | 0-1 | 0.2 |
| IgM++IgD+ | 11 | 0-6 | 0.4 |
| IgM++IgD− | 8 | 0-12 | 1.7 |
| IgG+ and IgA+ | 4 | 3-23 | 4.2 |
Number of cDNA clones.
Range of mutations per gene.
The PCR error rate of 0.15%16 was not subtracted from the values.
Most IgM-only PB B cells carry somatically mutated V region genes.The fraction of mutated cells among IgM-only B cells was determined by isolating single κ-positive IgM++IgD− PB B cells of a MACS-enriched B-lymphocyte fraction (Fig 1, right) using FACS and amplification of rearranged Vκ genes from the genomic DNA of the individual cells. In the first round of the PCR, a Jκ5 primer was used to prevent amplification of Vκ-Jκ1, 2, 3, and 4 products located away from the Cκ locus by inversion.29 In contrast to VκJκ rearrangements located in the Cκ locus, such rearranged genes may not be susceptible to somatic hypermutation due to their large distance from the κ intron-enhancer.30
In the single-cell PCR, controls consisted of single T cells isolated by FACS and reaction mixtures without cells. Water controls were always negative, whereas PCR products were obtained from some T cells. These were confirmed not to represent Vκ-Jκ rearrangements (data not shown). Of 16 IgM-only cells analyzed, 9 cells gave rise to a single and two cells to two distinct PCR products after second round amplification. PCR products were purified and directly sequenced. One product did not give rise to a readable sequence, probably because it represented a mixture of two rearrangements amplified from the corresponding cell. The sequences obtained from the remaining 12 products, all of which were unique, were compared with published Vκ sequences. Six sequences could be assigned to Vκ1 (2 to O2 and 1 each to A30, L12A, LFVk431 and O8) and three to Vκ2 (A19, A9, and A1) germline genes (Fig 2). One sequence showed the highest homology to the Vκ3 gene A27, and two sequences showed the highest homology to the single Vκ4 (B3) germline gene. Two cells (Mo11 and Mo33) carried both a functional and a nonfunctional rearrangement (Fig 2). Mo36 showed only an out-of-frame rearrangement. The Vκ-Jκ joints of the remaining cells were in-frame. Two rearrangements showed non–germline-encoded nucleotides at the Vκ-Jκ joints that presumably represent N region sequences inserted during Vκ-Jκ recombination (Fig 2).
Somatic mutation in Vκ genes amplified from IgM++IgD− PB B cells by single-cell PCR. (Upper part) Vκ sequences are designated Mo for IgM-only, followed by a sample number and the corresponding Vκ family to which the sequence could be assigned. Shown are the nucleotide differences in the Vκ sequences compared with the corresponding germline genes, which are indicated in the right column. The original references of the respective Vκ germline genes (with the exception of LFVk43131) are summarized in a recent review by Schäble and Zachau.27 LFVk431 might represent a polymorphic form of L1.31 Depicted are only those codons of the Vκ germline genes that differed from the respective Vκ sequences. CDRs and codons are numbered according to Kabat et al.32 (Dashes) Nucleotide identity to the corresponding germline genes. (Uppercase letters) Replacement mutations. (Lowercase letters) Silent mutations. (Dots) Codons are absent in the respective Vκ rearrangements. (Lower part) Nucleotide sequences of Vκ-Jκ junctions and number of nucleotide differences to the respective Vκ and Jκ33 germline genes. Indicated are the 3′end of the respective Vκ gene, N region nucleotides (N), and the 5′end of Jκ . (nf ) Nonfunctional rearrangement. The sum of nucleotide differences relative to the corresponding germline genes is shown in the right column. Mutations considered for the analysis are separated by a stretch of at least three identical nucleotides to the 3′Vκ and 5′Jκ ends, respectively. Underlined nucleotides as well as N-region insertions at the Vκ-Jκ junction were not counted as mutations. Sequences are available from EMBO/GenBank/DDBJ under accession no. Z82129-Z82167.
Somatic mutation in Vκ genes amplified from IgM++IgD− PB B cells by single-cell PCR. (Upper part) Vκ sequences are designated Mo for IgM-only, followed by a sample number and the corresponding Vκ family to which the sequence could be assigned. Shown are the nucleotide differences in the Vκ sequences compared with the corresponding germline genes, which are indicated in the right column. The original references of the respective Vκ germline genes (with the exception of LFVk43131) are summarized in a recent review by Schäble and Zachau.27 LFVk431 might represent a polymorphic form of L1.31 Depicted are only those codons of the Vκ germline genes that differed from the respective Vκ sequences. CDRs and codons are numbered according to Kabat et al.32 (Dashes) Nucleotide identity to the corresponding germline genes. (Uppercase letters) Replacement mutations. (Lowercase letters) Silent mutations. (Dots) Codons are absent in the respective Vκ rearrangements. (Lower part) Nucleotide sequences of Vκ-Jκ junctions and number of nucleotide differences to the respective Vκ and Jκ33 germline genes. Indicated are the 3′end of the respective Vκ gene, N region nucleotides (N), and the 5′end of Jκ . (nf ) Nonfunctional rearrangement. The sum of nucleotide differences relative to the corresponding germline genes is shown in the right column. Mutations considered for the analysis are separated by a stretch of at least three identical nucleotides to the 3′Vκ and 5′Jκ ends, respectively. Underlined nucleotides as well as N-region insertions at the Vκ-Jκ junction were not counted as mutations. Sequences are available from EMBO/GenBank/DDBJ under accession no. Z82129-Z82167.
Nine of the 10 cells analyzed harbored mutations in at least one of their rearranged Vκ genes (range, 2 to 13; Fig 2). From one cell, only an unmutated (out-of-frame) Vκ2 rearrangement was obtained. Nucleotide differences in the Vκ sequences relative to the corresponding Vκ and Jκ germline genes were considered for the determination of the somatic mutation frequency (Fig 2, bottom). The calculated value of 1.9% is similar to the values obtained for Vκ3 transcripts from IgM-only PB B cells of a 67-year-old16 and a 27-year-old donor (2.0% and 1.7%, respectively; Table 1). Also, both the fraction of mutated sequences among the respective sequence collections and the number of mutations per mutated V gene are in the same range.
As described in the following paragraph, a small fraction of IgM-only B cells (10 to 20%; Fig 3) express the CD5 antigen. This observation prompted us to determine the level of somatic mutation in CD5− and CD5+ IgM-only B cells by single-cell PCR. Whereas almost all CD5− IgM-only B cells were mutated, the vast majority of CD5+ IgM-only cells — like naive IgM+IgD+/CD5+ cells — showed no mutations in their rearranged Vκ genes, thus indicating that they represent a separate population of IgM-only cells (M. Fischer, U.K., K.R., and R.K., unpublished observations). Nevertheless, the single-cell analysis shows that the majority of IgM-only B cells carry mutated V genes.
Fluorescence analysis of CD5 and CD23 expression of human PB B-cell subsets. CD19+-enriched B cells (96% and 98% purity, respectively) were stained with anti-IgM-PE, IgD-CyChrome, and anti-CD5-FITC or anti-CD23-FITC. (Top) Windows were set around the IgM+/++IgD+, IgM++IgD−, and IgM−IgD− (mostly representing IgG+ and IgA+ cells) populations and analyzed for CD5 and CD23 staining within the respective fractions. (Histograms) CD5 and CD23 stainings of the CD19-enriched cell suspension (upper row) and the corresponding analyses of the B-cell subsets.
Fluorescence analysis of CD5 and CD23 expression of human PB B-cell subsets. CD19+-enriched B cells (96% and 98% purity, respectively) were stained with anti-IgM-PE, IgD-CyChrome, and anti-CD5-FITC or anti-CD23-FITC. (Top) Windows were set around the IgM+/++IgD+, IgM++IgD−, and IgM−IgD− (mostly representing IgG+ and IgA+ cells) populations and analyzed for CD5 and CD23 staining within the respective fractions. (Histograms) CD5 and CD23 stainings of the CD19-enriched cell suspension (upper row) and the corresponding analyses of the B-cell subsets.
IgM-only PB B lymphocytes are nonactivated, resting B cells resembling classical, class-switched memory cells but not naive or GC B cells.The existence of somatic mutations in IgV genes of IgM-only PB B cells suggests that they represent GC-derived memory B cells, like isotype-switched cells. However, the cells could also be GC B cells that have left the GC prematurely. Tonsillar GC B cells, in contrast to memory cells, express CD38, and some are also CD10+ and/or CD77+.28,34 In addition, at least 50% represent proliferating cells (as indicated by expression of Ki-6728 [and our own observations]). To examine whether IgM-only PB B cells show phenotypic features of GC B cells, B cell-enriched fractions from four donors were stained with anti-IgM-PE, anti-IgD-CyChrome, and the respective FITC-conjugated antibodies to CD38, CD10, and CD77 and analyzed on a FACScan. Like isotype-switched PB B cells, IgM-only cells were found to be negative for these surface antigens (data not shown). Furthermore, almost no cells were found to express the activation-antigens IL-2 receptor (CD25) or transferrin receptor (CD71; data not shown), in accordance with earlier reports on the low frequency of CD25+ and/or CD71+ B cells in the PB.35-37 We performed a cell cycle analysis to determine whether the IgM-only subset in the PB represents a resting or a proliferating population. IgM++IgD− and IgM+IgD+ B cells were isolated by FACS and subsequently analyzed for DNA content by quantitative propidium iodide staining.21 A proliferating IgM+ EBV cell line was analyzed in parallel. Whereas 20.5% of cells of the EBV line were in the S or G2/M phase of the cell cycle, the corresponding percentages of the IgM++IgD− and IgM+IgD+ cells were 0.4% and 0.5%, respectively (Fig 4). Thus, like class-switched PB B cells,37 IgM-only cells are resting.
Cell cycle status of a proliferating EBV line (left), IgM-only (middle), and IgM+IgD+ (right) B-cell subsets determined by propidium iodide staining.21 The percentages of cells in the S or G2/M phase of the cell cycle are indicated.
Cell cycle status of a proliferating EBV line (left), IgM-only (middle), and IgM+IgD+ (right) B-cell subsets determined by propidium iodide staining.21 The percentages of cells in the S or G2/M phase of the cell cycle are indicated.
CD5 is expressed on a variable percentage (10% to 20%) of tonsillar non-GC and adult PB B cells, most of which are IgD+.38 B-cell–enriched fractions from five donors were analyzed for CD5-expressing cells among the separate B-cell subsets (Fig 3). Whereas very few CD5+ B cells were found within the IgM−IgD− compartment, both IgM-only and IgM+IgD+ subsets contained similar amounts of CD5+ B cells (10% to 20%).
A surface antigen that appears to be differentially expressed on naive B cells on the one hand and class-switched memory B cells on the other is the IgE low-affinity receptor, CD23. This molecule is found on most IgD-positive PB B cells,39 whereas isotype-switched cells in both PB and tonsil do not express this antigen.34,37 39 We therefore analyzed IgM-only PB B cells of five donors for expression of CD23. Figure 3 shows the windows set for the analysis of CD23 expression of the respective subsets and the corresponding histograms from two experiments. Although anti-CD23 stainings on PB B cells derived from the various donors gave variable results with regard to the separation of CD23+ and CD23− subsets (see Fig 3), IgM-only cells were predominantly CD23−, like IgM−IgD− cells. On the other hand, most IgD+ cells were CD23+.
Taking these results together, IgM-only PB B cells phenotypically resemble classical memory cells rather than GC B cells and naive B cells.
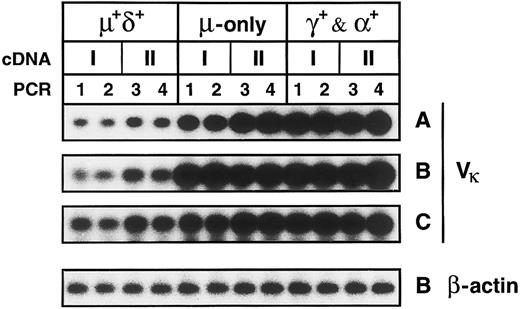
Somatically mutated IgM-only and class-switched memory B cells express higher levels of κ mRNA than naive B cells.IgM+IgD+, IgM++IgD−, and IgG+ and IgA+ PB B-cell subsets were isolated from three donors (A, B, and C; Fig 1). Two cDNA preparations from each of the respective fractions were amplified in separate PCR reactions using Vκ1-4 and Cκ and 5′ and 3′ β-actin primers. Gel-fractionated PCR products were blotted onto filters, and transfered products were hybridized with radioactively labeled Vκ or β-actin probes. The validity of the comparative PCR is discussed in the Materials and Methods. Figure 5 shows the blotted Vκ-Cκ PCR products as derived from the cellular subsets of the three donors. Bands were quantified using a phosphoimager. In each experiment, the average values determined for both the IgM++IgD− and IgG+ and IgA+ fractions were compared with the corresponding value of the IgM+IgD+ fraction; the latter value was set to 1. IgM-only cells expressed 6.7-, 6.7-, and 2.9-fold more κ mRNA, respectively, than naive B cells (Table 2). The κ mRNA levels in the IgG+ and IgA+ fraction were 11.2-, 8.0-, and 3.7-fold higher, respectively. On the other hand, the corresponding β-actin mRNA levels did not differ significantly: 1.3- to 1.6-fold and 1.5- to 1.9-fold higher in IgM-only and class-switched B cells, respectively, compared with naive B cells (Fig 5 and Table 2), indicating that the variation in the relative κ mRNA levels within a given subset is not due to loss of RNA during the isolation of the respective fractions.
Comparative PCR of cDNAs generated from PB B-cell subsets of three donors. Shown are bands corresponding to Vκ-Cκ PCR products derived from the PB B-cell subsets of donors A to C and the bands corresponding to β-actin PCR-products derived from the respective subsets of donor B. IgM+IgD+, IgM++IgD−, and IgG+ and IgA+ B-cell subsets and sample numbers of cDNA and PCR mixtures are indicated. Five thousand CE per cDNA mixture were amplified in separate reactions with Vκ-Cκ and β-actin primers.
Comparative PCR of cDNAs generated from PB B-cell subsets of three donors. Shown are bands corresponding to Vκ-Cκ PCR products derived from the PB B-cell subsets of donors A to C and the bands corresponding to β-actin PCR-products derived from the respective subsets of donor B. IgM+IgD+, IgM++IgD−, and IgG+ and IgA+ B-cell subsets and sample numbers of cDNA and PCR mixtures are indicated. Five thousand CE per cDNA mixture were amplified in separate reactions with Vκ-Cκ and β-actin primers.
κ Light Chain mRNA and (in Parentheses) β-Actin mRNA Levels of Human IgM-Only and Class-Switched PB B Cells Relative to Naive IgM+IgD+ PB B Cells
| Donor . | μ+δ+ . | μ++δ− . | γ+α+ . |
|---|---|---|---|
| A | 1.0 | 6.7 (1.6) | 11.2 (1.9) |
| B | 1.0 | 6.7 (1.3) | 8.0 (1.7) |
| C | 1.0 | 2.9 (1.3) | 3.7 (1.5) |
| Donor . | μ+δ+ . | μ++δ− . | γ+α+ . |
|---|---|---|---|
| A | 1.0 | 6.7 (1.6) | 11.2 (1.9) |
| B | 1.0 | 6.7 (1.3) | 8.0 (1.7) |
| C | 1.0 | 2.9 (1.3) | 3.7 (1.5) |
In vitro activated, proliferating B cells are known to express substantially higher levels of Ig mRNA than nonactivated, resting B cells, and plasma cells might express up to 100-fold more Ig mRNA transcripts than resting B cells (Kelley and Perry40 and references therein). Thus, elevated Ig mRNA levels in the IgM-only and class-switched subpopulations might be explained either by the occurrence of activated B cells and/or plasma cells within the subsets or by an increased Ig mRNA content in all or most cells of the respective subsets. The latter possibility is supported by the observations that, in vivo, the frequency of activated B cells rich in mRNA for Ig41 as well as Ig-secreting B cells14 among PB B lymphocytes is very low. Almost no activated and proliferating cells could be found within the IgM-only and IgG+ and IgA+ fractions (see previous section). Plasma cells were presumably eliminated during the magnetic cell separation, because they do not express CD19.42,43 In addition, because these cells carry little or no surface Ig,43 they are unlikely to contaminate the fractions that were sorted according to surface Ig expression. Nonetheless, to confirm the absence of cytoplasmic Ig-positive plasma cells in the sorted fractions, cytoplasmic stainings of cytospins generated from the respective B-cell subsets were performed. Using anti-κ-FITC/anti-λ-PE, no cytoplasmic Ig-positive cells could be detected among 3 × 104 cells (6 times more cells than used per reaction in the comparative PCR) of the IgM+IgD+, IgM-only, and IgG+ and IgA+ fractions (data not shown). Tew et al44 described plasmablasts present in the blood and lymph of the mouse that seem to be GC-derived precursors of not yet terminally differentiated plasma cells. If such plasmablasts exist in the human, they would probably also not account for the increased Ig mRNA levels in the IgM-only and IgG+ and IgA+ subsets. A recent study by Arpin et al45 suggests that human GC B cells differentiating into plasma cells, although downregulating pan-B cell markers such as CD20, retain surface CD38. Moreover, recent work characterized early plasma cells as CD38+, surface Ig-negative cells.43 As mentioned in the previous section, we did not observe CD38+ cells among the IgM-only and class-switched lymphocytes, nor did these subsets contain any cells otherwise phenotypically resembling GC B cells. Thus, it appears that elevated κ mRNA levels relative to those in naive IgM+IgD+ B cells are characteristic for most or all IgM-only and IgG+ and IgA+ cells.
DISCUSSION
Ig cDNA libraries generated from unseparated PB B cells are skewed towards somatically mutated transcripts derived from IgM-only and IgG+ and IgA+ memory B cells.The levels of κ mRNA were up to sevenfold and 11-fold higher in the IgM-only and class-switched fractions, respectively, than in the naive IgM+IgD+ subset (Table 2). Clearly, this must influence the analysis of the human PB B-cell repertoire using cDNA libraries generated from unseparated PB B lymphocytes. The elevated Ig mRNA levels in both IgM-only cells, which represent at the most 12% of IgM+ PB B cells16,19,20 and to which mutated Ig transcripts in IgM-expressing B cells are confined (see Table 1), and isotype switched B cells might explain the overrepresentation of somatically mutated transcripts in cDNA libraries generated from PB B cells.14,15,17,18,46,47 In such κ cDNA libraries, up to 90% of the transcripts were found to carry point mutations,46,47 although naive B cells (which express unmutated V genes) comprise more than 70% of all PB B lymphocytes.16,19,20 If, for example, the PB B-cell pool is composed of 75% IgM+IgD+, 10% IgM-only, and 15% class-switched B cells, the latter subsets expressing 7 and 11 times more κ mRNAs, respectively, than naive B cells, the fraction of somatically mutated Vκ transcripts in the corresponding cDNA library will be 76%. The large percentage of mutated κ transcripts in cDNA libraries generated from splenic B cells47 48 probably also reflects elevated Ig mRNA levels in somatically mutated splenic B-cell subsets rather than a higher proportion of cells carrying mutated V region genes.
IgM-only PB B cells seem to represent the IgM-expressing counterpart of classical class-switched memory B cells.IgM-only B cells of the PB resemble class-switched cells in many aspects. Most cells of both subsets express somatically mutated V region genes, albeit with a different load of mutations. They are neither in an activated state nor in cell cycle. They express higher κ mRNA levels than naive B cells. In contrast to naive B cells, they are mostly CD23−. In addition, both somatically mutated class-switched and IgM-only PB B cells are CD5− (M. Fischer, U.K., K.R., and R.K., unpublished observations).
Up to now, we analyzed rearranged Vκ genes (on both cDNA library and single cell levels) expressed by IgM-only PB B cells from four individuals differing from 27 to 67 years of age. Our collection of more than 40 sequences (Klein et al16; Table 1 and Fig 2; and M. Fischer, U.K., K.R., and R.K., unpublished observations) suggests that the somatic mutation frequency in IgM-only cells is characteristically about half that of class-switched cells, at least in the PB. That somatically mutated transcripts derived from IgM-expressing B cells show less mutation than transcripts derived from isotype-switched lymphocytes is supported by several studies analyzing μ-chain– as well as γ-chain–expressing B cells derived from either the tonsillar GC and memory fractions,34,49 the PB,17 or spleen.50 It should be noted that, in expressed Vk genes derived from both IgM-only and class-switched PB B cells, a pattern of nucleotide substitutions characteristic for somatic hypermutation as described by Neuberger and Milstein51 (eg, a clear preference for G to A transitions) could be observed (data not shown).
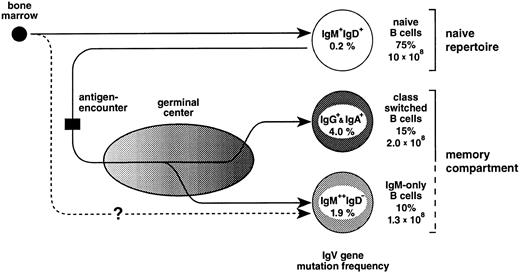
In the mouse, a fraction of peripheral (splenic) IgM-only cells represents recent emigrants from the bone marrow52 that express unmutated V region genes and have not yet acquired the IgM+IgD+ phenotype.53 The finding in the present work that the vast majority (Fig 2) of IgM-only PB B cells carry somatically mutated V genes indicates that, in humans, this subset is mainly composed of B cells at an advanced stage of cellular maturation (Fig 6). However, an intriguing alternative for the generation of mutated IgM-only PB B cells should also be considered. In the sheep, the primary antibody-repertoire is generated through somatic hypermutation in the absence of external antigens.54 This process takes place early in ontogeny within the ileal Peyer's patches.55 Perhaps IgM-only B cells, although resembling class-switched memory cells, are not GC-derived but represent naive B cells expressing V genes diversified by somatic mutation in, eg, gut-associated tissues resembling those of the sheep (Fig 6, lower pathway). As yet, there is no evidence for such structures in the human, and the hypothetical alternative pathway of IgM-only cells given in Fig 6 remains elusive. Indeed, as we had previously suggested,49 one observation strongly supports the notion that IgM-only PB B cells are GC-derived, namely that the somatic mutation frequencies of IgM-only PB and IgM-positive GC B cells (1.9% and 2.0%, respectively49; Fig 6) correspond to each other, like those of class-switched PB and GC B cells49 (4.0% and 3.3%, respectively; Fig 6).
The peripheral blood B-cell repertoire in humans and its presumptive generation. Naive B cells, the precursors of GC B cells, become activated by antigen and subsequently establish GCs. In the course of the GC reaction, class-switched and IgM-only B cells carrying antibody mutants are generated and, after selection, released into the periphery as memory B cells (upper pathway, see also text). The lower pathway represents a GC-independent generation of IgM-only cells. For the determination of mutation frequencies, sequences from previous studies16 and the present work (Table 1 and Fig 2) were considered. Indicated are also the percentages of the various subsets among PB B cells and total cell numbers in the PB of a normal adult (for calculation, see text).
The peripheral blood B-cell repertoire in humans and its presumptive generation. Naive B cells, the precursors of GC B cells, become activated by antigen and subsequently establish GCs. In the course of the GC reaction, class-switched and IgM-only B cells carrying antibody mutants are generated and, after selection, released into the periphery as memory B cells (upper pathway, see also text). The lower pathway represents a GC-independent generation of IgM-only cells. For the determination of mutation frequencies, sequences from previous studies16 and the present work (Table 1 and Fig 2) were considered. Indicated are also the percentages of the various subsets among PB B cells and total cell numbers in the PB of a normal adult (for calculation, see text).
Taken together, the memory B-cell compartment in the human PB seems to be composed of both isotype switched and IgM-expressing somatically mutated B cells. Support for this comes from two recent studies of T-cell–dependent immune responses to foreign antigens in humans. Bye et al56 generated heterohybridomas specific for the erythrocyte Rh(D) alloantigen from the PB (taken more than 2 weeks after boosting) of one individual and found the rearranged V genes of both IgM+ and IgG+ hybridomas (2 IgM and 3 IgG) to be somatically mutated. Oshiba et al57 analyzed the antibody response to TT and observed a considerable increase in the frequency of antigen-specific B cells in the PB of immunized individuals compared with that of unimmunized donors even years after immunization. Most of those B cells expressed IgM. We consider it likely that both the IgM+ B cells giving rise to the anti-Rh(D)–specific heterohybridomas56 and the TT-specific IgM+ cells57 represent GC-derived IgM-only memory cells.
To test directly whether TT-specific IgM-expressing memory B cells exist in the PB, we collaborated with C. Arpin and Y.-J. Liu at the Laboratory for Immunological Research (Dardilly, France) to culture IgM-only, class-switched, and naive B cells from PB of adults (immunized with TT within the last year) in the CD40-ligand system as described previously for tonsillar B-cell subsets.28 However, the outcome of the experiments led to the conclusion that the frequency of TT-specific B cells within either of the PB B-cell subsets cultured was too low to allow a quantification using this assay.
The IgM memory compartment in humans.Somatically mutated IgM-only B cells may exist in the various organs of the lymphoid system. The splenic marginal zone is largely composed of a B-cell subset that resembles IgM-only cells of the PB with regard to the expression of high levels of surface IgM but no or little IgD and the absence of CD23 (for review, see Kraal58 ). These cells by far outnumber class-switched cells located in the same area. A recent V gene analysis of marginal zone B cells scratched from tissue sections showed that the majority of rearranged V genes amplified from the genomic DNA of these cells harbored somatic mutations, indicating that most marginal zone IgM-expressing B cells are GC-derived.59 That the marginal zone contains memory B cells had been previously suggested by Liu et al60 and MacLennan et al61 based on studies in the rat. In human lymph nodes, the existence of B cells resembling marginal zone B cells (IgM++IgD−, CD23−), located at the outer area of the follicular mantle, has been noted.62 63
The occurrence of IgM-only B cells in the tonsil is indicated by the finding of mutated μ transcripts within the tonsillar memory B-cell fraction (IgD−CD38−CD23−) as defined by Pascual et al.34 Also, in the gut, B cells resembling IgM++IgD− cells of the PB and the marginal zone occur at a high frequency in the dome region of intestinal Peyer's patches.64,65 Expression of somatically mutated V genes of dome region B cells scratched from tissue sections has recently been shown.66
Based on the analysis of isotype distribution among human PB B cells16,19,20 (and this report), it appears that the PB on average contains about 75% naive, 15% class-switched, and 10% IgM-only B cells. On the assumption that the mean number of lymphocytes is 2.5 × 109 per 1l PB,67 of which 10% represent B cells,67 and that the human body contains 5 L of blood, the total number of PB B lymphocytes is 1.3 × 109, of which 1.3 × 108 represent IgM-only and around 2 × 108 represent class-switched cells. A normal, adult spleen comprises about 7 × 1010 lymphocytes,68 60% of which are B cells. Assuming that the distribution of the three B-cell subsets among those is roughly the same as in the PB, this lymphoid organ contains about 4 × 109 IgM-only and 6 × 109 class-switched cells. Because it is believed that the spleen comprises only 10% to 15% of the total B-cell pool,69 the total number of somatically mutated IgM-only and class-switched cells in the human lymphoid system is probably much higher. Thus, the IgM-only cells in the PB, in the splenic marginal zone, in tonsil, and perhaps also in lymph nodes and the intestine of humans presumably represent a large compartment of memory B cells.
Why do the IgV gene somatic mutation frequencies in IgM-only and class-switched cells differ by a factor of two? Based on the similar mutation frequencies in IgM-only PB B cells and IgM-expressing GC B cells, we have previously proposed that the former cells represent GC B cells that leave the GC at an early phase of the GC reaction without undergoing class-switching.49 Thus, in the course of a GC reaction, memory B cells would be constantly released into the periphery; and on the assumption that class-switch takes place predominantly in an advanced stage of the GC reaction, IgG+ and IgA+ cells would be expected to have undergone more rounds of somatic mutation than IgM-only cells (Fig 6). Alternatively, if the mutation frequency in IgM-only cells that is in between that of naive and class-switched cells reflects, on the average, an intermediate affinity to the respective antigen, this could also suggest that the affinity threshold during selection for improved antigen binding is lower in cells expressing the IgM as opposed to, eg, the IgG-receptor complex. Thus, IgM-only and class-switched GC B cells may have different signalling requirements, and the selection of a GC B cell into the memory compartment would depend on the class of the Ig-receptor complex expressed at this particular stage of differentiation. Still other possibilities would be that IgM-only and class-switched memory B cells have different precursors or that the quality or quantity of T-cell help determines whether a memory B lymphocyte undergoes class-switching or not. Be this as it may, a picture emerges in which the classical class-switched memory B-cell compartment is complemented, in humans, by an IgM memory compartment of almost equal size.
Recent studies by Toellner et al70 in the mouse point toward a possible fate of IgM-expressing memory B cells. Early after secondary immunization, antigen-specific B cells migrate from the splenic marginal zone (which is mainly populated by IgM-only B cells) to the T-zone and the red pulp, perform class-switching, and differentiate into plasma cells. This implies that IgM memory cells resident in the marginal zone, quickly after reencounter with specific antigen, can switch isotype and differentiate into plasma cells. Whether IgM-only memory B cells in addition can establish GCs is so far unresolved.
Malignancies of IgM-only B cells.Several B-cell malignancies are known in which the tumor consists of IgM+, IgD− B cells. In recent V gene analyses of four such malignancies, namely sporadic Burkitt's lymphoma,71,72 mucosa-associated lymphoid tissue (MALT) B-cell lymphoma,73,74 monocytoid B-cell lymphoma,75 and diffuse large-cell lymphoma,76 the rearranged V genes were found to be somatically mutated.
We had analyzed Vκ genes rearranged by sporadic Burkitt's lymphoma (7 IgM+, 1 IgG+) and found a somatic mutation frequency (1.8%) that is in a similar range as that characteristic for IgM-only PB and IgM+ GC B cells (1.9% and 2.0%, respectively), pointing to a relation with regard to developmental status between these lymphomas and IgM-only cells.71 Tamaru et al,72 also analyzing sporadic Burkitt's lymphoma, come to a similar conclusion. The somatic mutation frequencies of the other lymphomas mentioned above are mostly higher compared with those found in IgM-only cells, indicating that the corresponding cells underwent more rounds of somatic mutation. Monocytoid B-cell lymphomas share similarities with distinct B cells located at the sinusoidal structures in lymph nodes. It has been proposed that those monocytoid B cells represent the lymph node equivalent of marginal zone B cells of the spleen.77 The malignant cells of MALT B-cell lymphomas also phenotypically resemble splenic marginal zone IgM-only cells.73 74
The similarities between those tumors and IgM-only B cells suggests that these malignancies originate from neoplastic transformation of IgM-expressing germinal center B cells or their descendants, namely IgM-only memory B cells.
ACKNOWLEDGMENT
We thank C. Göttlinger for help with the FACS, W. Müller and U. Betz for assistance in the densitometric analysis, and U. Ringeisen for the graphical work. We are grateful to C. Arpin and Y.-J. Liu (Laboratory for Immunological Research, Schering-Plough, Dardilly, France) for sharing unpublished data. We also thank J. Irsch (Institute for Genetics, Cologne, Germany) for many helpful discussions and for providing the EBV line 2C5E7.
Supported by the Deutsche Forschungsgemeinschaft through Di184 and Sonderforschungsbereich 243.
Address reprint requests to Ulf Klein, Institute for Genetics, University of Cologne, Weyertal 121, 50931 Cologne, Germany.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal