CYTOKINE RECEPTORS are cell-surface glycoproteins that bind specifically to cytokines and transduce their signals. These receptors enable cells to communicate with the extracellular environment by responding to signals generated in the vicinity or in other parts of the organism. Thus, the initial binding of cytokines to their receptors is a key event that occurs rapidly, at very low cytokine concentrations, is usually virtually irreversible, and leads to intracellular changes resulting in a biologic response. The biologic response can vary between cytokine receptors and from cell to cell but in general it involves gene expression, changes in the cell cycle, and release of mediators such as cytokines themselves.
Cytokine receptors function as oligomeric complexes consisting of typically two to four receptor chains that may be the same or different. In single subunit receptors the subunits fulfill the dual role of binding to cytokines and signaling. Examples of receptors that use a single type of subunit are those for growth hormone (GH), erythropoietin (EPO), granulocyte colony-stimulating factor (G-CSF ), and thrombopoietin (TPO). In multi-subunit receptors the different subunits may perform specialized functions such as ligand-binding or signal transduction. Multi-subunit receptors may consist of two subunit types such as the receptors for granulocyte-macrophage CSF (GM-CSF ), interleukin-3 (IL-3), and IL-5 where an α subunit is specific for each ligand and a β subunit is common to all three (βc ), with both chains participating in signaling. The IL-6 receptor also contains two subunit types, IL-6Rα and gp130. However, in this case the function of each chain is more exclusive, with IL-6Rα being the major binding protein with no direct role in signaling, and gp130 being the signal transducer. Receptors that contain three different subunits are the CNTF receptor (CNTFR), formed by the CNTFRα chain, gp130, and the leukemia inhibitory factor (LIF ) receptor, and the IL-2 receptor (IL-2R) which consists of the IL-2Rα chain or tac (which is not a typical member of the cytokine receptor family), IL-2Rβ, and IL-2Rγ, with the latter two being the signaling molecules.
The cloning of cytokine receptors has shown a striking structural and functional conservation which has justified their distinct grouping into the cytokine receptor superfamily. However, it is becoming clear that within this superfamily, structurally similar subfamilies exist whereby some receptors or receptor subunits are more related to each other than to other members of the receptor superfamily. For example the recently cloned receptor for TPO (TPOR) is more closely related to the EPO receptor (EPOR) and βc than to other cytokine receptors.
In functional terms some receptors have subunits that subserve similar functions. For example, the common β subunit (βc ) shared by the GM-CSF, IL-3, and IL-5 receptors is functionally analogous to gp130, which is the common subunit of the IL-6, CNTF, cardiotrophin, oncostatin M, LIF, and IL-11 receptors, to IL-2Rγ which is shared by the receptors for IL-2, IL-4, IL-7, IL-9, and IL-15, and to the common subunit of the IL-4 and IL-13 receptors.1 2 These common subunits have the dual function of affinity-converting the initial cytokine binding into a high-affinity state, and of being the major signal transducer in each of these receptor systems. The communal nature of these subunits helps explain much of the overlapping activities of the different cytokines in each receptor system.
This review focuses mostly on these communal subunits and, in particular, on the structure of the common β subunit (βc ) of the human GM-CSF, IL-3, and IL-5 receptors and the mechanism of activation of this receptor family. This review does not address the activation pathways and signaling molecules associated with βc following receptor activation, topics covered by recent reviews elsewhere.3-5 This review discusses recent modeling, mutagenesis, and functional studies on βc likely to provide a paradigm on which predictions on other communal subunits may be based. In particular, the identification of regions important for the binding of GM-CSF, IL-3, and IL-5 may offer a novel strategy to interfere with the function of several cytokines at once at the cell surface. Evidence is also reviewed and presented that subtle differences in the way the communal subunits associate with the major binding subunits may exist, which has implications for the general mechanism of receptor activation and the biologic function of cytokines.
Sequences of the CRM From Various Signaling Subunits of Cytokine Receptors Are Compared
| CRM: . | βc1 . | βc2 . | LIFR1 . | LIFR2 . | gp130 . | TPOR1 . | TPOR2 . | EPOR . | IL2Rβ . | GHR . | PRLR . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| βc1 | 18.0 | 12.0 | 17.0 | 17.7 | 14.9 | 16.1 | 15.7 | 17.6 | 14.7 | 16.2 | |
| βc2 | 23.2 | 13.7 | 18.4 | 20.8 | 19.8 | 19.1 | 23.1 | 16.3 | 15.3 | 14.0 | |
| LIFR1 | 14.7 | 16.2 | 20.3 | 17.7 | 14.0 | 15.6 | 18.4 | 12.4 | 12.6 | 21.6 | |
| LIFR2 | 15.4 | 16.6 | 19.6 | 29.5 | 11.7 | 15.0 | 18.8 | 16.3 | 13.6 | 18.4 | |
| gp130 | 16.8 | 22.7 | 18.6 | 21.3 | 15.4 | 17.7 | 17.4 | 14.7 | 17.9 | 24.9 | |
| TPOR1 | 16.6 | 16.8 | 14.9 | 12.8 | 16.4 | 19.5 | 28.2 | 14.9 | 18.6 | 21.1 | |
| TPOR2 | 15.7 | 17.5 | 17.2 | 16.9 | 18.8 | 17.2 | 21.3 | 17.4 | 15.5 | 19.7 | |
| EPOR | 15.6 | 20.4 | 15.7 | 18.6 | 19.4 | 26.6 | 20.7 | 18.5 | 17.9 | 19.6 | |
| IL2Rβ | 17.4 | 20.5 | 11.9 | 16.3 | 14.4 | 17.7 | 15.5 | 16.8 | 17.2 | 16.9 | |
| GHR | 15.7 | 18.5 | 13.7 | 15.6 | 21.7 | 21.3 | 17.5 | 18.1 | 14.4 | 32.0 | |
| PRLR | 19.1 | 18.6 | 19.8 | 20.3 | 27.5 | 23.2 | 17.6 | 20.3 | 17.0 | 31.5 |
| CRM: . | βc1 . | βc2 . | LIFR1 . | LIFR2 . | gp130 . | TPOR1 . | TPOR2 . | EPOR . | IL2Rβ . | GHR . | PRLR . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| βc1 | 18.0 | 12.0 | 17.0 | 17.7 | 14.9 | 16.1 | 15.7 | 17.6 | 14.7 | 16.2 | |
| βc2 | 23.2 | 13.7 | 18.4 | 20.8 | 19.8 | 19.1 | 23.1 | 16.3 | 15.3 | 14.0 | |
| LIFR1 | 14.7 | 16.2 | 20.3 | 17.7 | 14.0 | 15.6 | 18.4 | 12.4 | 12.6 | 21.6 | |
| LIFR2 | 15.4 | 16.6 | 19.6 | 29.5 | 11.7 | 15.0 | 18.8 | 16.3 | 13.6 | 18.4 | |
| gp130 | 16.8 | 22.7 | 18.6 | 21.3 | 15.4 | 17.7 | 17.4 | 14.7 | 17.9 | 24.9 | |
| TPOR1 | 16.6 | 16.8 | 14.9 | 12.8 | 16.4 | 19.5 | 28.2 | 14.9 | 18.6 | 21.1 | |
| TPOR2 | 15.7 | 17.5 | 17.2 | 16.9 | 18.8 | 17.2 | 21.3 | 17.4 | 15.5 | 19.7 | |
| EPOR | 15.6 | 20.4 | 15.7 | 18.6 | 19.4 | 26.6 | 20.7 | 18.5 | 17.9 | 19.6 | |
| IL2Rβ | 17.4 | 20.5 | 11.9 | 16.3 | 14.4 | 17.7 | 15.5 | 16.8 | 17.2 | 16.9 | |
| GHR | 15.7 | 18.5 | 13.7 | 15.6 | 21.7 | 21.3 | 17.5 | 18.1 | 14.4 | 32.0 | |
| PRLR | 19.1 | 18.6 | 19.8 | 20.3 | 27.5 | 23.2 | 17.6 | 20.3 | 17.0 | 31.5 |
GENE ORGANIZATION OF βc AND OTHER CYTOKINE RECEPTOR SUBUNITS
Molecular cloning of the cDNA encoding human βc6 predicts the protein to be an 880-amino acid molecule with four 100-amino acid extracellular domains related to fibronectin type III domains, a single membrane-spanning sequence and some 450 intracellular residues. The four extracellular domains are comprised of seven β strands (denoted A-G) and are organized in two cytokine receptor modules (CRM). Subsequent analysis of the gene structure of βc and that of related receptors, and comparison with protein structures based on the paradigm of the GH receptor (GHR) suggest a relationship between gene organization and functional regions in the receptor.
Conservation of both position and phase of intron/exon boundaries has been noted in cytokine receptor genes.7 The organization of the genes encoding the mouse β chains, the communal β chain AIC2B, and the IL-3–specific β chain AIC2A have been determined.8 As with other genes from the cytokine receptor superfamily, each 100 amino acid fibronectin type III domain is encoded by two exons. The intron/exon boundaries that delineate the ends of each domain are of the phase 1 type in which the intron disrupts the codon after the first nucleotide. The intervening intron/exon boundaries for domains one and three of AIC2A and AIC2B are of type 2 where the intron disrupts the codon after the second nucleotide, and type 0 for domains two and four with the intron interrupting the reading frame between codons. This pattern of intron/exon boundaries, conserved both in position and the phase of the introns, has been described as the 1-2-1-0-1 rule.7 As with other cytokine receptor genes, the transmembrane region and immediate cytoplasmic portion in the genes for AIC2A and AIC2B are encoded in two small exons. This is followed by a nonconserved cytoplasmic exon and a large exon encoding the C-terminus.
The gene organization of βc differs from that of the prototypic GHR in two respects. Firstly, there is a direct duplication of the CRM, similar to that seen in the TPOR. Secondly, the region encoding the C-terminus is interrupted by an additional phase 2 intron, a feature also seen in the IL-4R,9 although the cytoplasmic regions of these proteins do not exhibit any especial similarity. The two CRM of βc apparently arose as a result of a duplication because they are somewhat more closely related to each other than to other cytokine receptors (Table 1). This is similar to the leptin receptor (OBR), in which the two CRM are more similar to each other than to other receptors, but differs from other receptors such as the TPOR in which the N-terminal CRM is more closely related to the EPOR than to the C-terminal CRM, and the LIFR in which the second CRM is more closely related to gp130 than to the first CRM.
The conservation of intron/exon boundaries in the cytokine receptor family suggests that they may delineate structural or functional regions in the proteins. This is clearly the case for the boundaries between extracellular domains of the GHR, where the intron separates sequences encoding the two pairs of cysteine residues that form intramolecular disulfides and lie immediately C-terminal to the C strand. However, in GHR, the intron in the second domain interrupts the D strand. Sequence alignment10 and molecular modeling11 suggest that the introns of βc also occur between the C and D strands in the first and third domains and interrupt the D strands in the second and fourth domains. The conserved cytoplasmic exon has been found to encode sequences essential for signal transduction by βc12 and similar regions have been identified in GM-CSF receptor α chain (GMRα),12 IL-3 receptor α chain (IL-3Rα),13 the IL-5 receptor α chain (IL-5Rα),14 and IL-2R β chain (IL-2Rβ).15
PROTEIN STRUCTURAL FEATURES OF CYTOKINE RECEPTORS
The sequences of the hematopoietic cytokine receptors exhibit a conserved region of approximately 200 amino acid residues (cytokine receptor module, CRM) that was proposed to consist of two β barrel structural domains.16 Several sequence motifs may be recognized as typical of this family of receptors. Each domain contains a conserved Trp near its N-terminus, the first domain contains four conserved cysteine residues thought to be involved in disulfide bonds, while the second domain has two or three Pro residues near its N-terminus, an alternating pattern of hydrophobic residues (YXVXVRVR consensus) and an especially well-conserved WSXWS motif near its C-terminus. The elucidation of the structure of the GHR complexed with GH17 has provided some explanation of the role of these conserved residues. Each domain of the GHR consists of two β-pleated sheets containing the A, B, and E strands and the D, C, F, and G strands, respectively. The general topology of these strands can be seen in the βc model (Fig 1). The conserved N-terminal Trp residues lie in the central, B, strand of the first sheet and constitute part of the hydrophobic cores of the proteins. These residues interact with conserved hydrophobic residues from the central C and F strands of the opposing sheets including the YXVXVRVR motif in the second domain. The WSXWS motif exists as a β bulge facilitating interdigitation of the two Trp residues between the surface-orientated Arg side-chains of the YXVXVRVR motif in the F strand. Although the GHR has Tyr and Phe residues in place of the more usual Trp in the WSXWS motif, the aromatic rings of these residues superimpose on the Trp heterocycles in the structure of the closely related prolactin receptor.18 The elucidation of the structure of the GH/GHR complex has provided a structural paradigm on which our understanding of the structure and function of other cytokine receptors can be built.
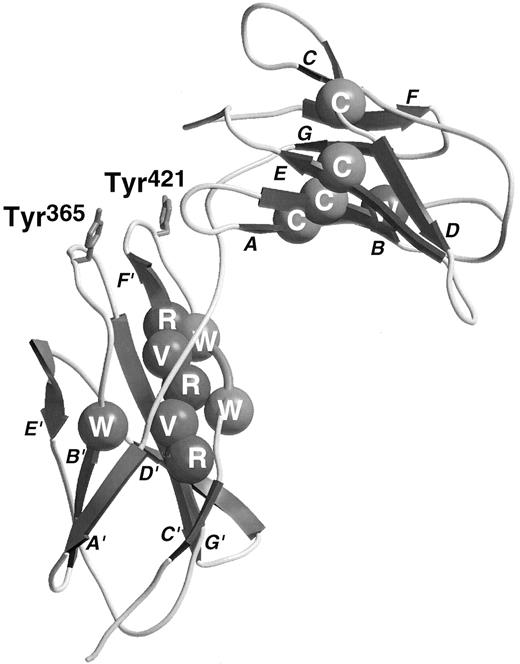
Ribbon diagram of the third and fourth domains of βc with conserved residues shown by CPK spheres. Conserved residues are colored as follows: Cys, yellow; buried hydrophobics, green; Arg, blue; Trp of WSXWS sequence, purple (see cover figure). The two Tyr residues involved in ligand-binding are drawn in red stick form (see cover figure). The strands are labeled in close proximity to the arrow illustrating the direction of the relevant strand.
Ribbon diagram of the third and fourth domains of βc with conserved residues shown by CPK spheres. Conserved residues are colored as follows: Cys, yellow; buried hydrophobics, green; Arg, blue; Trp of WSXWS sequence, purple (see cover figure). The two Tyr residues involved in ligand-binding are drawn in red stick form (see cover figure). The strands are labeled in close proximity to the arrow illustrating the direction of the relevant strand.
Within the CRMs, each structural domain is related to the archetypal fibronectin type III domain but they are not closely related to each other. Although the amino acid sequences are highly divergent, this common cytokine receptor structural motif has probably been conserved throughout mammalian evolution and has been found in other vertebrates such as birds.19 This conservation is illustrated by, firstly, the presence of sequence motifs such as the WSXWS motifs, disulfide pairs, and several other residues (eg, Trp in B strand of both domains). Secondly, in the vast majority of cytokine receptors, the cytokine receptor domains are present as pairs constituting a CRM. In addition to CRM, several receptors have recruited additional domains into their extracellular regions such as classical type III fibronectin domains, Ig domains, or cytokine-receptor second domains (IL-3, IL-5, GM-CSF receptor α chains). Although the intracellular portions of the cytokine receptors are highly variable, a membrane-proximal region of approximately 50 amino acids typically contains proline motifs and is encoded by an exon of conserved structure. This region has been implicated in signal transduction via association with members of the JAK family of protein tyrosine kinases.20 21
PROTEIN STRUCTURE OF βc
Extracellular regions.The extracellular domains of βc bear sufficient resemblance to those of GHR to permit molecular modeling by homology. Lyne et al modeled the membrane-proximal CRM of human βc permitting the prediction of contact sites for the ligands and receptor α-chains. The model supported the notion22,23 that the B′-C′ and F′-G′ loops in the fourth domain of βc are involved in ligand recognition and also proposed that, like the GH receptor, the E-F loop of the third domain would be involved in ligand-binding. The major role of Tyr residues in the domain 4 B′-C′ and F′-G′ loops that have been identified by mutagenesis (vide infra) was not highlighted in the models of the IL-3 and GM-CSF receptor complexes. Of interest with respect to receptor activation process is the observation of the formation of disulfide-linked IL-3Rα/β receptor heterodimers on binding of IL-3 leading to functional activation of these β-chains.24 The N-terminal domain of βc has seven cysteine residues and the second domain has one. The first two cysteine residues are encoded by one exon and probably form a conserved disulfide between the A and B strands. A second disulfide between the D and E strands is also predicted to occur leaving three nonconserved cysteine residues, two of which (Cys86 and Cys91 ) lie in the C-D loop where they may either form an additional disulfide or be available for intermolecular disulfide formation. The remaining cysteine residues (100 and 234) are not present in the mouse β chains.
Cytoplasmic regions.Nothing is known of the tertiary structure of the cytoplasmic domains of members of the cytokine receptor family. However, in addition to their extracellular homology, several signaling subunits of cytokine receptor systems exhibit additional sequence similarities in the membrane-proximal cytoplasmic regions denoted “box 1” and “box 2”25 (Fig 2). The region described as box 1 contains a basic motif (LRR), a Trp residue followed by proline-containing sequences (IPNP). In βc confirmation of the importance of box 1 came from deletion analysis which defined a region comprising residues 456 to 487, encompassing box 1, as being critical for mediating growth response when transfected into Ba/F3 cells.12 Sequences corresponding to box 1 are found in a number of cytokine receptors where they probably serve a common function in the recruitment of kinases of the JAK family. In addition to box 1 residues, a region of βc comprising residues 517 to 542, that includes box 2 sequences, was found to be required for the full sensitivity of the biologic response.12 Sequences corresponding to box 2 exhibit a weak consensus of PXXLE followed by several charged residues and are encoded near the 5′ of the final large exon of the signaling subunits of receptors for which the gene structures are known at a distance of typically 16 to 24 residues C-terminal to a conserved Trp residue (Fig 2). In βc , this distance is apparently greater (39 residues), probably due to the interruption of the final exon by the third cytoplasmic intron.
Sequences of the immediate cytoplasmic regions of various human receptor signaling subunits. The positions of intron/exon boundaries are indicated by (▿). In some cases, these have been inferred from the mouse genes. Conserved residues are boxed with a consensus indicated below; aliphatic (f ), hydrophobic (Ω), hydrophilic (⋄), or the conserved residue.
Sequences of the immediate cytoplasmic regions of various human receptor signaling subunits. The positions of intron/exon boundaries are indicated by (▿). In some cases, these have been inferred from the mouse genes. Conserved residues are boxed with a consensus indicated below; aliphatic (f ), hydrophobic (Ω), hydrophilic (⋄), or the conserved residue.
These regions have been noted to be involved in the function of related receptors as well. For example, in the β chain of the IL-2R, a 13-amino acid deletion of the box 1 region resulted in a 50-fold decrease in the ability to signal in a transient proliferation assay.15 Furthermore, box 2 in IL-2Rβ is essential for the spectrum of activities stimulated by IL-2.15 Determination of the functional relationship between boxes 1 and 2 requires determination of the structure of this immediate cytoplasmic region.
The cytoplasmic region of βc also contains several tyrosines that are phosphorylated after cytokine binding. A major one is Tyr750, substitution of which abolishes phosphorylation of βc ,26 which is associated with impaired viability. Tyr577, on the other hand, is essential for Shc phosphorylation.27,28 The role of phosphorylated receptor tyrosines in recruiting associated proteins and linking receptors to cellular functions is discussed elsewhere,4 but a picture is emerging whereby tyrosine-based motifs can be identified in signaling receptor subunits that couple the receptor to specific substrates.29
STRUCTURAL REQUIREMENTS FOR βc FUNCTION
Regions in βc involved in ligand binding.As described above, the extracellular portion of βc is comprised of two CRM. Most other receptor subunits of the cytokine receptor superfamily including the α chains for GM-CSF, IL-3, and IL-5 contain only a single copy of the cytokine receptor module. The role of the individual cytokine receptor domains of βc is poorly understood. Studies to identify ligand binding determinants have focussed on the membrane proximal CRM, which is thought to be better positioned for interactions with ligand and α chain.
The βc is thought to be involved in direct interaction with each of its ligands in the high-affinity receptor complex. Evidence for this comes from structure-function studies on the ligands themselves. Substitution of a conserved glutamate in the first α helix of GM-CSF,30-32 IL-3,33 or IL-534 abolishes high-affinity binding without affecting the ability of the mutated cytokines to bind to their specific α chains. Despite the complete loss of high-affinity binding by all the glutamate-substituted mutants, the net effect on biologic activity is not all or none, but dependent on the amino acid used to substitute the conserved glutamate. The most clear example is GM-CSF where substitution of Glu21 for different amino acids leads to analogues of varying potency, and in the case of a charge reversal (E21R, E21K), to analogues devoid of classical GM-CSF activity that behave as antagonists.31,35 This spectrum of biologic activities by “low affinity only binding” GM-CSF mutants suggests that residual and weak interactions with βc , not detectable by binding experiments, are taking place. Alternatively, the Glu21 mutants may conformationally affect receptor dimerization and the formation of high-order GM-CSF receptor complexes required for receptor activation (see below). As βc acts as an affinity converter in the receptor complex these findings suggest that the conserved glutamate motif is involved in βc interaction. This is supported by cross-linking studies showing that the ligand is capable of being cross-linked to βc , indicating that ligand and βc are in close proximity and suggesting that the interaction between the conserved glutamate in the α helix of GM-CSF, IL-3 or IL-5, and βc may be a direct one.
From the crystal structure of the GH-GHR complex, three regions of GHR were found to be solvent inaccessible in the ligand bound receptor complex and, therefore, represent contact sites between the ligand and the receptor.17 These regions lie in the surface exposed intervening regions between the β strands of the GHR CRM; in the E-F, B′-C′ and F′-G′ loops. Mutagenesis in these regions has shown important similarities and differences within the cytokine receptor superfamily.
The E-F loop has been shown to be involved in ligand binding in several cytokine receptors. Mutation studies on GHR identified Trp104 in the E-F loop as being critical for interaction with GH.36 No detectable binding occurred when this residue was substituted with alanine, whereas a more conservative substitution with phenylalanine reduced affinity for GH 110-fold. In the IL-5Rα, Arg188 in the putative E-F loop was shown to be involved in IL-5 binding.37 Alanine substitution of this residue resulted in more than a 100-fold loss in affinity for IL-5. Also, recently a residue in the E-F loop of EPOR, Phe97, was identified by mutagenesis studies as being important for EPO binding.38 Substitution of Phe97 to alanine resulted in a 1,000-fold reduction in EPO binding affinity whereas substitution with tyrosine or tryptophan had a much less dramatic effect, suggesting that an aromatic hydrophobic residue is required for high-affinity EPO binding.
In contrast, no effect on high-affinity GM-CSF or IL-3 binding was observed by penta-alanine substitution of the PVPDP sequence predicted to lie in the E-F loop of βc (unpublished observations, 1996), suggesting that this region is not important for ligand binding. However, the primary sequence similarity of βc with GHR in this region is extremely weak, raising the possibility that the region targeted for mutagenesis was not analogous to the E-F loop of GHR. Further studies in βc may yet reveal a role for this region in ligand interaction.
A second region in GHR, the B′-C′ loop, was identified as being involved in interaction with GH.39 Mutagenesis studies identified Trp169 in the B′-C′ loop as being a key hydrophobic residue required for ligand binding. Previously, the predicted B′-C′ loops of IL-6Rα,40 IL-2Rβ,41 and AIC2A42 were implicated as being involved in ligand binding. Intriguingly, as with GHR, the residues identified in each case include a large aromatic hydrophobic residue. This suggests, as has been found with Trp104 and Trp169 in the GH-GHR complex, that hydrophobic interactions are important for ligand binding in many cytokine receptors.
In βc , the putative B′-C′ loop was targeted for mutagenesis in two independent studies.22,43 In both studies His367 was identified as being important for high-affinity GM-CSF binding. Our study also showed that not only His367 but also the hydrophobic residues Tyr365 and Ile368 were important for IL-5 as well as GM-CSF high-affinity binding, but individually had only a minor role in IL-3 high-affinity binding.22 These findings demonstrated that GM-CSF, IL-3, and IL-5 may interact with βc in different ways and suggested that it may be possible to develop antagonists against βc that are selective in their effect on different ligands. This observation is in parallel to results obtained with mutant ligands, where charge reversal substitution of the conserved glutamate residue in the first α helix of GM-CSF (Glu21 ) and IL-5 (Glu13 ) facing βc44 not only completely abolished the biologic activity of the cytokines but also rendered them functional antagonists. Similarly, mutation of IL-3 at Glu22 severely disrupted the biologic activity of the cytokine but, in this case, the analogues still functioned as agonists, suggesting that residual functional interactions were occurring with βc .33
In the IL-6Rα, residues in the F′-G′ loop were identified as being important for ligand binding and an aromatic hydrophobic residue, Phe298 was implicated.40 In the crystal complex of GH-GHR, residues of the F′-G′ loop were judged to be solvent inaccessible, suggesting an interaction between this loop of the receptor and the ligand.17 However, mutagenesis studies showed that there was no productive interaction between GH and this loop in the receptor.39 This is in direct contrast to the role of the F′-G′ loop in βc in which a single residue, Tyr421, has been identified as playing a key role in high-affinity binding of all three ligands.23 Indeed, functional studies showed a loss in IL-3–stimulated activation of STAT5 greater than 10,000-fold. Significantly, a mutant βc in which the entire putative F′-G′ loop with the exception of Tyr421 was substituted with alanine was able to support high-affinity GM-CSF and IL-3 binding, indicating that this residue alone in the context of the putative loop region was sufficient for binding.23 Alignment of the predicted F′-G′ loops of other members of the cytokine receptor family reveals the presence of hydrophobic aromatic residues in several cytokine receptors.23 This suggests that this region may also be involved in ligand binding in other receptor systems and may represent an ideal target for small molecule antagonists. In particular, in the case of βc , it may be envisaged that compounds targeting Tyr421 could simultaneously inhibit the actions of GM-CSF, IL-3, and IL-5. These molecules may have therapeutic benefit in diseases such as asthma where all three cytokines have been implicated. Further support for the global importance of this region in βc and in other receptors23 stems from the observation that a small EPO mimetic peptide competitively binds to this region in the EPO receptor.45 46
The involvement of hydrophobic residues in ligand binding sites has become a recurrent theme in mutagenesis studies to date in members of the cytokine receptor family. In GHR, 11 residues make up the functional binding epitope and form a hydrophobic core in which Trp104 and Trp169 form the major interaction with GH that accounts for more than 75% of the binding free energy.39 Also, the residues involved in ligand interaction in GHR lie in the intervening loop regions between β strands that are flexible and in close proximity to the ligand. Thus, the conservation of structural features in the cytokine receptor family maintains the structural organization of the receptor subunits so that ligand binding sites in the loops are effectively presented for efficient ligand interaction.
In terms of communal receptor subunits that interact with multiple ligands, the βc may provide a paradigm for ligand interaction. It is clear that in βc there are ligand-specific and shared interaction sites for GM-CSF, IL-3, and IL-5. It would be interesting to examine whether similar observations can be made with gp130, LIFRβ, and the IL-2Rβ chain.
Regions important for structural integrity.Several studies have examined the effect of mutating the conserved structural determinants of cytokine receptors on ligand binding. Paired cysteine residues are a conserved feature in the membrane distal domain of cytokine receptors. In studies on IL-6R,40 the prolactin receptor,47 and GMRα chain,48 it has been shown that mutation of these cysteines disrupted ligand binding, suggesting that these cysteine residues define the secondary structure of the receptor. In addition, these cysteines may also be involved in disulfide interactions between βc and the IL-3Rα chain.24 It is anticipated that mutagenesis of cysteine residues in the membrane distal CRM of βc will show those residues involved in this intermolecular interaction and also determine those involved in intramolecular interactions.
A tryptophan in the B′ strand is a conserved feature of the cytokine receptor superfamily. In the GHR crystal structure this residue forms part of the hydrophobic core of the membrane proximal domain.17 Substitution of this residue (Trp358 ) in βc completely abrogates high-affinity GM-CSF binding and IL-3–induced receptor activation,49 indicating that maintenance of the hydrophobic core is crucial for receptor integrity and consequently ligand interaction.
The role of the highly conserved WSXWS motif in cytokine receptors has long been the subject of controversy. Studies have focused on this motif in several different cytokine receptors including IL-2Rβ,50 EPOR,51,52 IL-6R,40 GHR,53 the GMR α chain,48,54 and IL-3R α chain.13 Mutations in this motif were found to have various effects on cell-surface expression of these receptors, ligand binding, and receptor internalization, although the structural basis for these effects was not understood. However, a recent systematic study of mutants in EPO receptor has more clearly defined the structural role of the WSXWS motif.55 One hundred WSXWS mutants representing all the single amino acid substitutions for each of the five residues of the motif were analyzed for their effect on cell-surface expression, ligand binding, and functional response. Only conservative amino acid substitutions of the tryptophan and serine residues were tolerated, although the effect of mutation was on the secretory pathway, reducing the ability of the receptor polypeptide to exit the endoplasmic reticulum and, consequently, cell-surface expression was affected. However, the affinity and functional activity of the mutant EPO receptors that reached the cell surface was unchanged, indicating that the ligand binding site was unaltered by these mutations.
From the crystal structure of the growth hormone receptor, the WSXWS motif forms a close interaction with another conserved motif, VRXR. The two motifs bind the F′-G′ loop and interact to form a hydrophilic-aromatic stack, which thereby constrains the F′-G′ loop. One of the inconsistencies with WSXWS mutations in different receptors has been their effect on ligand binding affinity. This may be as a consequence of the disruption of the F′-G′ loop, which makes a major contribution to ligand interaction in some receptors, as in the βc , but not in other receptors, as in GHR. To date no WSXWS studies on βc have been published, but on the basis of the involvement of the F′-G′ loop in ligand binding we would predict that should WSXWS mutants of βc be cell-surface expressed, there would be a dramatic effect on ligand binding.
MECHANISM OF CYTOKINE RECEPTOR ACTIVATION
Once cytokines bind to surface receptors they induce receptor clustering or oligomerization followed by receptor activation and the generation of intracellular signals. Ligand-dependent receptor dimerization or oligomerization appears to be a general feature of cytokine receptors,56 which applies to members of the cytokine receptor superfamily as well as to members of the receptor tyrosine kinase family. Receptor dimerization/oligomerization can be seen in cases where the receptors are constituted by single chains as with c-kit,57 the GH receptor,58 EPOR,59 and G-CSFR,60 as well as in multi-subunit receptors such as the IL-661 and IL-324 receptors. Receptor dimerization is more often than not induced by ligand. In the cytokine receptor superfamily, GH has been shown to induce GHR dimerization,58 and IL-6 and IL-3 also induce dimerization of their respective receptors.24,61 On the other hand, EPO does not seem to play a role in dimerizing the EPOR.59 An intermediate example is the G-CSFR where G-CSF induces the conversion of a dimeric receptor into a tetrameric one.60
In the GM-CSF, IL-3, IL-5 receptor subfamily, intriguing differences are beginning to emerge that may have implications for the role of these receptors in hematopoietic cell function. The major difference lies in the absolute requirement for IL-3 and IL-5 for dimerization of IL-3Rα with βc , and IL-5Rα with βc whereas, in contrast, at least some of the GM-CSFR probably exists as a preformed complex. In some experiments GM-CSF has been shown to facilitate the co-immunoprecipitation of GMRα with βc62,63; however, other experiments using mutated receptors and mutated GM-CSF analogues suggest the existence of a preformed GMRα-βc heterodimeric complex. In the case of a mutated GMRα where the second conserved cysteine was replaced, GM-CSF was unable to bind to this receptor alone, but bound with high affinity when this mutant receptor was coexpressed with βc .30 Reciprocally, a mutated GM-CSF molecule carrying an Asp112 mutation showed no detectable binding to GMRα alone, yet exhibited nearly full wild-type activity in cells expressing both GMRα and βc .30 Both cases suggest that the presence of βc in a preformed complex with GMRα compensates for losses in the GM-CSF:GMRα binding. The intrinsic interaction of GMRα and βc appears to be weak (with GM-CSF stabilizing the GMRα:βc complex by 1,000-fold),64 but nevertheless of sufficient strength to be detectable in cells coexpressing soluble GMRα and membrane-bound βc using anti-subunit–specific monoclonal antibodies.65 Using a similar cell-based system we have found that in stably transfected cells expressing full-length GMRα or full-length IL-3Rα together with soluble βc , antibodies to βc detect surface staining on the former but not on the latter cells (unpublished data, 1996), supporting the notion that βc association with GMRα and IL-3Rα is fundamentally different.
It is possible that GMRα and βc are selectively coexpressed and cotransported to the endoplasmic reticulum and the cell surface. In structural terms it is possible that E′ strands and A-B loops in domain four of βc and domain 2 in GMRα form a favorable interface allowing subunit-subunit interaction akin to the GH receptor 1 and GH receptor 2 interaction. The biologic significance of a preformed GM-CSFR complex is unclear but may facilitate a small amount of signal to trickle into the cell. It is worth noting the ubiquitous presence of the GM-CSFR in hematopoietic cells and the pivotal role it plays in functions such as survival66,67 and apoptosis.68,69 It is also interesting to note that cell survival is, of all the functions triggered by GM-CSF, the one that requires the smallest amount of GM-CSF,70 71 indicating that very low levels of receptor occupancy and presumably receptor dimers are required for signaling survival. It is possible that a preformed GM-CSFR complex affords a small and transient amount of “survival signaling” that gives the cell “time” to encounter the appropriate cytokine in the immediate environment.
Disulfide-linked receptor complexes in the GM-CSF, IL-3 receptor family. (A) Immunoprecipitation of 125I-surface-labeled primary leukemic cells with anti-βc monoclonal antibody in the absence of ligand or in the presence of IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under nonreducing or reducing (4% 2-mercaptoethanol [2-ME]) conditions. (B) Immunoprecipitation of MO7e cells with anti-βc monoclonal antibody and immunoblotting with antiphosphotyrosine antibody. The cells were either untreated or treated with GM-CSF or IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under non-reducing or reducing (4% 2-ME) conditions.
Disulfide-linked receptor complexes in the GM-CSF, IL-3 receptor family. (A) Immunoprecipitation of 125I-surface-labeled primary leukemic cells with anti-βc monoclonal antibody in the absence of ligand or in the presence of IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under nonreducing or reducing (4% 2-mercaptoethanol [2-ME]) conditions. (B) Immunoprecipitation of MO7e cells with anti-βc monoclonal antibody and immunoblotting with antiphosphotyrosine antibody. The cells were either untreated or treated with GM-CSF or IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under non-reducing or reducing (4% 2-ME) conditions.
The mechanism of receptor dimerization involves either two identical receptor subunits (homodimerization) or two different subunits of the receptor (heterodimerization). Examples of the former are the single-chain receptors. The GH, G-CSF, and EPO receptors have been shown to homodimerize followed by recruitment of the appropriate tyrosine kinase and signaling. The receptor for TPO (TPOR) would also be expected to undergo homodimerization. In multi-subunit receptors such as the IL-6 and IL-3 receptors, both heterodimerization and homodimerization have been shown to occur. In the case of the IL-6R, IL-6 triggers the heterodimerization of IL-6Rα, the major binding subunit, with gp130, the signaling subunit.25 This in turn allows the homodimerization of gp130 to a second gp130 molecule.72 In the case of the related CNTF receptor, CNTF heterodimerizes with gp130 while gp130 also heterodimerizes with the LIFR to initiate signaling.73 In the case of the IL-3R, IL-3 triggers heterodimerization of IL-3Rα with βc .24 In the GM-CSF, IL-3, IL-5 receptor family, homodimerization of βc24,63 (Fig 3) as well as heterodimerization of the GMRα and βc (unpublished results, 1996) have been observed, both cases in the absence of stimulus. Stimulation with GM-CSF, on the other hand, leads to further heterodimerization of GMRα with βc62 in a similar fashion to IL-3–induced heterodimerization of its IL-3Rα with βc . Whether ligand induces further homodimerization of βc is not yet clear.
Dimerization of cytokine receptor subunits has been shown to occur by covalent and noncovalent means. The dimerization of single-chain receptors has been shown to be noncovalent such as with the GHR17 but also covalent involving disulfide bonds as with the EPOR.74 With the IL-6R and CNTFR the heterodimerization of the ligand binding, nonsignaling α subunits to gp130 is noncovalent; however, the association between the signaling subunits involves disulfide linkage.72,73 Dimerization of both the GM-CSF and IL-3 receptors is different to the receptors mentioned above in that a disulfide-linkage has been observed between each of the binding α subunits and βc .24 Figure 3A illustrates this point by showing that in the presence of IL-3 two high-molecular-weight complexes are induced. These have been shown to be disulfide-linked and to contain IL-3Rα and βc .24 In the absence of IL-3 a 240,000 molecular-weight band is seen representing a disulfide-linked βc homodimer.
In the IL-6, LIF, CNTF and EPO receptors, nondisulfide as well as disulfide-linked receptor dimers are associated with receptor activation as measured by phosphotyrosine reactivity of the dimers. Similarly, in the case of the GM-CSFR, both disulfide and nondisulfide-linked dimers are tyrosine phosphorylated upon the addition of GM-CSF (Fig 3). In contrast, in the case of the IL-3R, the majority of the phosphotyrosine reactivity is associated with the disulfide-linked complex and very little, if any, with the nondisulfide complexes24 (Fig 3), indicating that the formation of the former is essential for receptor activation. This concept is further supported by the demonstration that prevention of this disulfide-linked association between IL-3Rα and βc by iodoacetamide inhibits receptor phosphorylation.24 Interestingly, IL-3 high-affinity binding is not prevented by iodoacetamide, indicating that high-affinity binding and receptor phosphorylation are dissociable events.24 These experiments and the presence of IL-3 in only the noncovalently linked α-β heterodimers24 suggest a sequence of events in which IL-3 binds initially to IL-3Rα and the IL-3:IL-3Rα associates with βc through CRM2, forming a high-affinity noncovalently-linked complex. The bringing together of IL-3Rα and βc may facilitate the disulfide linkage of a free cysteine in domain 1 of βc with a free cysteine in the N-terminal domain of IL-3Rα. A similar interaction may occur with the unpaired Cys in the N-termini of GMRα and IL-5Rα. The resulting effect is receptor activation by phosphorylation. A recurrent theme in receptor activation is that the major receptor signaling subunits, not only βc but also gp130 and LIFRβ in the IL-6 and CNTF receptor systems, become phosphorylated. In contrast, none of the α subunits show evidence of phosphorylation.
These experiments illustrate a fundamental difference between the IL-6R and the GM-CSF, IL-3, IL-5 receptor families in terms of the contribution of the receptor α chains. Thus, while the IL-6Rα chain does not form disulfide-linked dimers with gp130 and its cytoplasmic portion is not essential for receptor activation, the GMR, IL-3R, and IL-5R α chains have been shown to require the cytoplasmic domain of the respective receptor α chains,12-14, 75 and at least IL-3Rα and GMRα form disulfide-linked complexes (Fig 3).
It is interesting to note that in addition to GMRα or IL-3Rα covalent heterodimerization with βc , recent experiments have also noted that βc homodimerization is sufficient for receptor activation. This has been shown using chimeric molecules expressing extracellular GMRα chain domains and cytoplasmic βc domains. Cells expressing these chimeras together with wild-type βc can proliferate in the presence of GM-CSF.76 This is analogous to the constitutive activation of the EPOR by an extracellular R129C mutation which leads to receptor dimerization.77 Similarly, chimeric EPOR/βc with the R129C mutation causes constitutive JAK-1/JAK-2 phosphorylation and factor-independent growth,78 indicating that homodimerization of βc is sufficient for signaling. Although this induced βc homodimerization (mediated by either ligand or a mutant Cys-Cys linkage) leads to receptor activation, we have also observed the existence of spontaneous covalently linked βc homodimers in the absence of ligand in primary cells (Fig 3A). However, these βc dimers are not phosphorylated (Fig 3B) and phosphotyrosine reactivity is observed only when ligand is added to the cells. Similarly, βc dimers were demonstrated by cross-linking experiments that were phosphorylated in response to ligand.63 This suggests that under normal conditions the α chain is required to activate the receptor through both the initial binding of ligand and by facilitating βc dimerization. The existence of disulfide-linked βc homodimers in the absence of ligand, to which disulfide-linked heterodimers can be added by the presence of ligand, may favor the formation of hexameric complexes necessary for receptor activation (see below) analogous to the IL-6R system. Hexameric IL-6:IL-6R complexes have already been shown by analytical centrifugation79 and IL-6 mutagenesis and IL-6R immunoprecipitation studies.80 This stoichiometry is facilitated by the ability of IL-6 to interact with its receptor through three distinct binding sites, one for IL-6Rα and two for gp130. The ability of gp130 to homodimerize completes and stabilizes the hexamer.
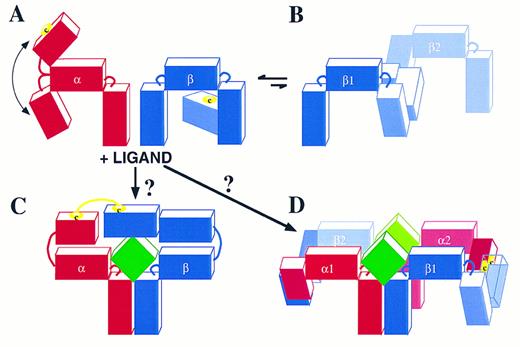
In the case of the GM-CSF, IL-3, IL-5 receptors, the stoichiometry of the ligand-receptor complex has not yet been defined. This system appears to be different from the IL-6R system in that so far ligand has been found to bind its receptor through two distinct sites; one comprising helix D30,81 and probably also helix C82 that interacts with the specific receptor α chains, and a second involving mainly the conserved Glu motif in Helix A30-32,83 that interacts with the B′-C′ and F′-G′ loops of domain 4 in βc .22,23,43 The formation of disulfide-linked α-βc heterodimers is critical in this ligand receptor complex24 (Fig 3). These are likely to occur through free cysteines present in the N-termini of each α subunit and a free cysteine present in domain 1 of βc . It is worth noting that the N-termini of the GMRα, IL-3Rα, and IL-5Rα subunits are not classical cytokine receptor modules and are not present in other members of the cytokine receptor superfamily. Furthermore, an uneven number of cysteines are present in each of the three α subunits, highlighting the availability of at least one cysteine for intermolecular bonding. From molecular modeling of βc the free cysteine in βc is likely to be at position 86 or 91 in domain 1. The remaining cysteines are in conserved positions which from cytokine receptor sequence alignment and modeling are likely to be important in intramolecular bonding and maintenance of structural integrity. Figure 4 shows the α and βc subunits of this class of cytokine receptors highlighting the putative free cysteine and the predicted approximate 80° angles which link each domain to the next. Of note is the flexibility of the linkage of the N-terminal domain of the α chain to the CRM, which would allow it to flex toward or away from the membrane (Fig 4A); however, it would be extremely unlikely to permit an angle of more than 125°. The IL-3Rα subunit and probably also the IL-5Rα subunit are not associated with βc in the absence of ligand. However, the GMRα subunit and βc are probably pre-associated through interaction of their membrane proximal domains. In the absence of ligand βc homodimerizes probably by disulfide linkage of its free cysteines (Fig 4B); however, this is a nonproductive interaction because no phosphorylation of βc is seen (Fig 3).63 The presence of ligand may induce complexes formed by trimers or hexamers. The formation of a trimer (Fig 4C) seems unlikely as it would require the hinge between the N-terminus and the CRM of the α subunits to exhibit an unusual flexibility to allow the α and βc subunits to form a disulfide bridge. Thus, a trimer in which ligand contacts both receptor subunits which in turn disulfide-link to each other seems unlikely because an angle of about 180° would be required (Fig 4C). Instead, we favor the possibility that, after ligand binding, two disulfides are formed, one involving an α subunit of receptor 1 with a βc subunit of receptor 2, and a second disulfide involving the βc subunit of receptor 1 with the α subunit of receptor 2 (Fig 4D). This model is consistent with the possible angles between receptor domains, the existence of high-order molecular-weight complexes by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)24 and the formation, under certain conditions, of ligand dimers themselves.84 85 Ultimately, analytical centrifugation and crystallization of the ligand-receptor complex may resolve this issue, but it is nevertheless apparent that the GM-CSF, IL-3, IL-5 receptor system offers a new type of structural assembly and dynamics of receptor subunit interaction. It remains to be seen whether the lessons learned from this system will apply to other members of the cytokine receptor superfamily.
Schematic representation of possible GM-CSF, IL-3, and IL-5 receptor complexes. (A) The receptor α and βc interaction in the absence of ligand. The cartoon illustrates that the N-terminus of the α chain has approximately 90° angle flexibility from the CRM. In the case of the IL-3R24 and probably also of the IL-5R, the α and βc subunits are not associated either covalently (disulfide linked) or noncovalently. In the case of the GM-CSFR, there is no disulfide linkage but there is probably a noncovalent association through domain 2 of the GMRα CRM and domain 4 of βc . (B) In a dynamic and probably reversible manner, βc can homodimerize by disulfide linkage. However, this is a nonproductive interaction because no tyrosine phosphorylation of βc is seen (see also Fig 3).63 (C) The binding of ligand to these receptors may then give rise to a trimer with a ligand:α:βc stoichiometry of 1:1:1. However, the 180° angle required for the α subunit to link up with βc makes this possibility unlikely. Instead, (D) illustrates a more likely ligand:α:βc stoichiometry of 2:2:2 where the α subunit of receptor 1 forms a disulfide bond with βc of receptor 2 and the α subunit of receptor 2 forms a disulfide bond with βc of receptor 1. The free cysteines in the α and βc subunits are illustrated.
Schematic representation of possible GM-CSF, IL-3, and IL-5 receptor complexes. (A) The receptor α and βc interaction in the absence of ligand. The cartoon illustrates that the N-terminus of the α chain has approximately 90° angle flexibility from the CRM. In the case of the IL-3R24 and probably also of the IL-5R, the α and βc subunits are not associated either covalently (disulfide linked) or noncovalently. In the case of the GM-CSFR, there is no disulfide linkage but there is probably a noncovalent association through domain 2 of the GMRα CRM and domain 4 of βc . (B) In a dynamic and probably reversible manner, βc can homodimerize by disulfide linkage. However, this is a nonproductive interaction because no tyrosine phosphorylation of βc is seen (see also Fig 3).63 (C) The binding of ligand to these receptors may then give rise to a trimer with a ligand:α:βc stoichiometry of 1:1:1. However, the 180° angle required for the α subunit to link up with βc makes this possibility unlikely. Instead, (D) illustrates a more likely ligand:α:βc stoichiometry of 2:2:2 where the α subunit of receptor 1 forms a disulfide bond with βc of receptor 2 and the α subunit of receptor 2 forms a disulfide bond with βc of receptor 1. The free cysteines in the α and βc subunits are illustrated.
Supported by grants from the National Health and Medical Research Council of Australia and the Anti-Cancer Foundation of the Universities of South Australia. C.J.B. is a Rotary Peter Nelson Leukaemia Research Fellow of the Anti-Cancer Foundation of the Universities of South Australia.
Address reprint requests to Angel F. Lopez, MD, PhD, Hanson Centre for Cancer Research, the Institute of Medical and Veterinary Science, Frome Rd, Adelaide 5000, Australia.


![Fig. 3. Disulfide-linked receptor complexes in the GM-CSF, IL-3 receptor family. (A) Immunoprecipitation of 125I-surface-labeled primary leukemic cells with anti-βc monoclonal antibody in the absence of ligand or in the presence of IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under nonreducing or reducing (4% 2-mercaptoethanol [2-ME]) conditions. (B) Immunoprecipitation of MO7e cells with anti-βc monoclonal antibody and immunoblotting with antiphosphotyrosine antibody. The cells were either untreated or treated with GM-CSF or IL-3. Proteins were separated on a 7.5% SDS-PAGE gel under non-reducing or reducing (4% 2-ME) conditions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/5/10.1182_blood.v89.5.1471/3/m_bl_0050f3.jpeg?Expires=1767727679&Signature=VnOeZruffBWXl3b0ubqJG5Vi2hkE9hiBMCGJY8eGbUD8Jfq~PUJ~vT8oDmotKfj2HsSDDU1fq~hnFe6CgrEeqEX5M18m9bi~g8OUY9XmURRvUK0CFIq3UlksoBTAYqsPRZRBq8fxNa94UthTjWAm0SVZiGHOZFSXf89LLH15fccUFWE4t8czdmz5~sGNh~60gSZa5WQJHbjQdfQ1HxWwVko2WwAKJ6ELgTmvhwl2T5lK0ppaGTWwnIWAleGWfn2SJboO9w-AE54ggu-tBl~Efr5R-T1NnVfg-X1fm915ihtiB~XZJPQsWC0kfIdEQYVvv5Dia22sDK7WGr5NfnMbFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal