Abstract
Translocation t(5; 12)(q33; p13), resulting in an ETV6/PDGFRB gene fusion, is a recurrent chromosomal abnormality associated with chronic myelomonocytic leukemia (CMML). An analogous translocation was also found in four cell lines with features of pre-B acute lymphoblastic leukemia (ALL). Using fluorescence in situ hybridization (FISH) we show here that in three of these cell lines identical complex rearrangements occurred. However, the regions involved on 5q and 12p are different from the breakpoints in CMML, and the translocation is accompanied by seemingly identical cryptic deletions of both 5q and 12p chromosome sequences in all analyzed pre-B ALL cell lines. The similar cytogenetic, FISH, and immunophenotyping findings in the three cell lines suggest that the t(5; 12)(q31q33; p12) defines a new entity of pre-B ALL.
TRANSLOCATION t(5; 12)(q33; p13) is a recurrent chromosomal abnormality in a subgroup of myeloid malignancies showing features of both myeloproliferative disorders (MPD) and myelodysplastic syndromes (MDS).1 The translocation results in a fusion of ETV6 (previously known as TEL ), which belongs to the ETS gene family of putative transcription factors, at 12p13, with PDGFRB mapped to 5q33.2 Recently, other chromosomal translocations involving ETV6 and different partners on chromosomes 3, 10, 22 were reported in myeloid disorders and on chromosomes 9 and 21 in lymphoid malignant disorders showing that ETV6 rearrangements are not lineage-restricted.3 Of particular importance is the t(12; 21) fusing ETV6 to CBFA2 (AML1 ), shown to be frequently associated with a deletion of the second ETV6 allele. This translocation defines a distinct entity of childhood pre-B acute lymphoblastic leukemia (ALL) and is the most frequent, but cytogenetically largely undetected, chromosomal anomaly in childhood ALL.
Translocation (5; 12) was also observed in some leukemic cell lines with features of pre-B lymphoblasts. These cell lines, NALM-6, PBEI, and SUP-B26/-B28, were recently listed together with several other leukemia-lymphoma cell lines characterized by specific chromosomal translocations and gene fusions by Drexler et al.4 Based on an analogous translocation occurring in chronic myelomonocytic leukemia (CMML), the investigators suggested that the t(5; 12) translocation in these pre-B ALL cell lines also involved ETV6 and PDGFRB. However, no molecular evidence supporting this hypothesis was provided.
We report here the characterization by fluorescence in situ hybridization (FISH) of the t(5; 12) translocation found in NALM-6, PBEI, and in a third cell line, LR-10.6, obtained from a B-ALL case (J.I.-E. and A.A., unpublished observation, July 1996). Our results indicate that this translocation is molecularly different from the t(5; 12) observed in CMML and consistently affects distinct loci both on 12p12 and on 5q31q33. In addition, FISH analysis showed that the t(5; 12) is invariably associated with two apparently identical cryptic deletions within the 5q31q33 as well as the 12p12 breakpoint regions of all three cell lines.
MATERIALS AND METHODS
Cell lines.NALM-6 was established in 1976 as one of several cell lines developed from the peripheral blood (PB) of a 19-year-old man who at that time had a first relapse of a non-T, non-B ALL.5,6 The PBEI cell line was established from a bone marrow (BM) culture of freshly isolated pre-B ALL cells by Pirrucello et al.7 The LR-10.6 cell line was established in 1993 from BM cells from a 5-year-old boy with B-ALL diagnosed in 1991, clinically in complete remission (<5% of blast cells) (J.I.-E., unpublished results, July 1996). Based on hematologic data (Table 1), all three cell lines seem to be arrested at an early stage in B-cell development and are considered to be of the pre-B phenotype. Translocation t(5; 12)(q31-q33; p12), previously described as 5q− and 12p+ in the case of NALM-6,6 was found in all cell lines. Occurrence of the translocation in the original patient's material was documented in two cases, namely NALM-6 and PBEI.
Results of Immunophenotypic and Cytogenetic Analysis
| Cell Line . | Immunophenotype (%) . | Karyotype* . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| . | Tdt . | CD19 . | CD10† . | Smlg . | cμ . | FMC7 . | HLA-DR . | CD24 . | . |
| NALM-6 | 40 | 45‡ | 70 | 1 | 38 | 1 | 70 | 0 | 46, XY, t(5; 12)(q33; p12) [10] |
| PBE-1 | 40 | 32‡ | 48 | 1 | 60 | 0 | 83 | 53 | 46, XY, t(5; 12)(q33; p12) [21] |
| 47, XY, t(1; 10)(q21; q22), t(5; 12)(q33; p12), + 13 [5] | |||||||||
| LR-10.6 | 90 | 99 | 99 | 14 | 100 | 1 | 100 | 9 | 46, XY, t(5; 12)(q33; p12) [20] |
| Cell Line . | Immunophenotype (%) . | Karyotype* . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| . | Tdt . | CD19 . | CD10† . | Smlg . | cμ . | FMC7 . | HLA-DR . | CD24 . | . |
| NALM-6 | 40 | 45‡ | 70 | 1 | 38 | 1 | 70 | 0 | 46, XY, t(5; 12)(q33; p12) [10] |
| PBE-1 | 40 | 32‡ | 48 | 1 | 60 | 0 | 83 | 53 | 46, XY, t(5; 12)(q33; p12) [21] |
| 47, XY, t(1; 10)(q21; q22), t(5; 12)(q33; p12), + 13 [5] | |||||||||
| LR-10.6 | 90 | 99 | 99 | 14 | 100 | 1 | 100 | 9 | 46, XY, t(5; 12)(q33; p12) [20] |
T-cell markers CD2, CD4, and CD8, myeloid markers CD13 and CD33, and megakaryocytic marker CD41 were consistently negative in all three cell lines.
Abbreviations: Tdt, terminal deoxynucleotidyl transferase; Smlg, surface membrane Ig; cμ, cytoplasmic μ chain.
In square brackets: number of cells analyzed.
Combined CD19/CD10 staining.
Weak staining.
FISH.FISH was performed as previously described8 using 11 biotinylated cosmid clones and two BAC clones hybridizing along the short arm of chromosome 12 and 13 cosmid or YAC probes specific for the 5q31-q35 region, listed in Table 2. The ETV6 locus at 12p13 was investigated with cosmids c179A6 and c148B6 containing, respectively, exon 1 or exon 8 of ETV6.9 Cosmid clones for GDI.D4 (c106B9), D12S1094 (c214F3), and D12S308 (c167H1) were isolated from the LL12NC01 library10 using polymerase chain reaction (PCR)-generated probes and standard procedures. The identity of the clones was confirmed by PCR. BAC clones for D12S308 (b204K22) and D12S1274 (b322023) were isolated from a commercial BAC library (Research Genetics, Huntsville, AL). Their identity was confirmed by PCR, and the insert length was measured by pulsed field gel electrophoresis (PFGE) to be 100 kb. Eight YACs assigned to the 5q31q33 region were selected from the sequence tagged site (STS)-based map constructed at the Whitehead Institute (MIT, Cambridge, MA).11 The ninth YAC used for FISH, y745D10, was a chimeric probe hybridizing to 2p21 and 5q33 and positive for PDGFRB.2 A genomic cosmid probe and a cDNA clone for CSF1R were obtained from A. Roebroek (Center for Human Genetics, Leuven, Belgium), whereas cosmids c33G8 (IL3), cosB (PDGFRB ), and cCI5-20 were kindly provided by M. Lovett (University of Texas Southwestern Medical Center at Dallas), M. Roberts (M.D. Anderson Cancer Center, The University of Texas, Houston), and Y. Nakamura (Cancer Institute, Tokyo, Japan), respectively. Chromosomes 12 were identified by cohybridization with a centromeric probe (pBR12, D12Z3) labeled with Texas Red-5-dCTP. All FISH results were collected on a Leitz DMRB fluorescence microscope (E. Leitz Inc, Wetzlar, Germany) equipped with a cooled black and white CCD camera (Photometrics, USA, Tucson, AZ) run by SmartCapture software (Vysis, Stuttgart, Germany). Ten to twenty metaphase cells were analyzed in each experiment.
Results of FISH Analysis
| Probes* . | Loci . | Localization . | Chromosomes . | |||
|---|---|---|---|---|---|---|
| . | . | . | 5 . | der(5) . | 12 . | der(12) . |
| c449A | D12S158 | 12p13.3 | − | + | + | − |
| c179A6 | ETV6, exon 1 | 12p13.1 | − | + | + | − |
| c148B6 | ETV6, exon 8 | 12p13.1 | − | + | + | − |
| c12pC6 | D12S178 | 12p12 | − | + | + | − |
| c214F3 | D12S1094 | 12p12 | − | + | + | − |
| b322O23 | D12S1274 | 12p12 | − | − | + | − |
| c167H1 | D12S308 | 12p12 | − | − | + | − |
| b204K22 | D12S308 | 12p12 | − | − | + | − |
| c106B9 | GDI.D4 | 12p12 | − | − | + | + |
| c1C3 | D12S119 | 12p12 | − | − | + | + |
| c144B6 | D12S932 | 12p12 | − | − | + | + |
| c242H11 | D12S930 | 12p12 | − | − | + | + |
| c185C6 | KRAS2 | 12p12 | − | − | + | + |
| c33G8 | IL3 | 5q31 | + | + | − | − |
| y773D3 | D5S396; -399; -89; IL9 | 5q31 | + | + | − | − |
| y773B7 | D5S1972; -1840; -1863, -1871 | 5q31 | + | − | − | − |
| y939F12 | D5S402; -436; -643; -210; -546; -547; -548; -549; -68; -686 | 5q31 | + | − | − | − |
| y888A7 | D5S638, -376 | 5q31 | + | − | − | − |
| y745D10 | CSF1R; D5S353; -551 | 5q33 | + | − | − | − |
| cCML182 | CSF1R | 5q33 | + | − | − | − |
| cosB | PDGFRB | 5q33 | + | − | − | − |
| y756A2 | D5S470, RPS14 | 5q33 | + | − | − | + |
| y816D6 | RPS14, SPARC, D5S519 | 5q33 | + | − | − | + |
| y748D10 | D5S673, -410, -670 | 5q33 | + | − | − | + |
| y913F6 | D5S1439, -487; -662; -820 | 5q33 | + | − | − | + |
| cC5l-20 | 5q34q35.1 | + | − | − | + | |
| Probes* . | Loci . | Localization . | Chromosomes . | |||
|---|---|---|---|---|---|---|
| . | . | . | 5 . | der(5) . | 12 . | der(12) . |
| c449A | D12S158 | 12p13.3 | − | + | + | − |
| c179A6 | ETV6, exon 1 | 12p13.1 | − | + | + | − |
| c148B6 | ETV6, exon 8 | 12p13.1 | − | + | + | − |
| c12pC6 | D12S178 | 12p12 | − | + | + | − |
| c214F3 | D12S1094 | 12p12 | − | + | + | − |
| b322O23 | D12S1274 | 12p12 | − | − | + | − |
| c167H1 | D12S308 | 12p12 | − | − | + | − |
| b204K22 | D12S308 | 12p12 | − | − | + | − |
| c106B9 | GDI.D4 | 12p12 | − | − | + | + |
| c1C3 | D12S119 | 12p12 | − | − | + | + |
| c144B6 | D12S932 | 12p12 | − | − | + | + |
| c242H11 | D12S930 | 12p12 | − | − | + | + |
| c185C6 | KRAS2 | 12p12 | − | − | + | + |
| c33G8 | IL3 | 5q31 | + | + | − | − |
| y773D3 | D5S396; -399; -89; IL9 | 5q31 | + | + | − | − |
| y773B7 | D5S1972; -1840; -1863, -1871 | 5q31 | + | − | − | − |
| y939F12 | D5S402; -436; -643; -210; -546; -547; -548; -549; -68; -686 | 5q31 | + | − | − | − |
| y888A7 | D5S638, -376 | 5q31 | + | − | − | − |
| y745D10 | CSF1R; D5S353; -551 | 5q33 | + | − | − | − |
| cCML182 | CSF1R | 5q33 | + | − | − | − |
| cosB | PDGFRB | 5q33 | + | − | − | − |
| y756A2 | D5S470, RPS14 | 5q33 | + | − | − | + |
| y816D6 | RPS14, SPARC, D5S519 | 5q33 | + | − | − | + |
| y748D10 | D5S673, -410, -670 | 5q33 | + | − | − | + |
| y913F6 | D5S1439, -487; -662; -820 | 5q33 | + | − | − | + |
| cC5l-20 | 5q34q35.1 | + | − | − | + | |
c, cosmid; b, BAC; y, YAC.
RESULTS
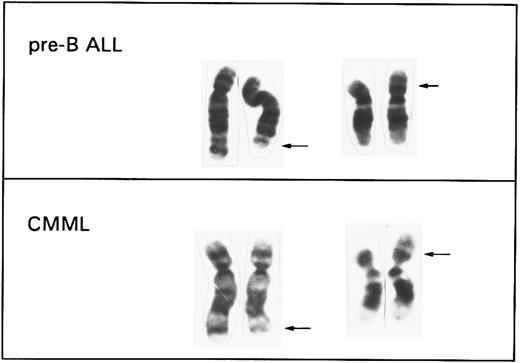
Two previously reported pre-B ALL cell lines, NALM-6 and PBEI, and a third one, LR-10.6, obtained by two of us (J.I.-E. and A.A., unpublished results, July 1996), were collected for a FISH study of the t(5; 12). Cytogenetic and immunophenotyping analysis of these three cell lines, performed at the time of FISH investigation and summarized in Table 1, confirmed the previously published data. A similar pattern of antigen expression, namely combined CD19/CD10 positivity, TdT positivity, cytoplasmic μ chain positivity, and negative SmIg staining in all three cell lines, indicates that they are indeed pre-B cells. The t(5; 12) was found in all cases in 100% of the cells analyzed. Additional chromosomal abnormalities, namely a t(1; 10)(q21; q22) and +13, that were identified in PBEI, represent secondary changes acquired during culturing of this cell line.7 A representative partial karyotype of a t(5; 12) from the cell line NALM-6 is shown in Fig 1, together with a t(5; 12) typically found in CMML.3
Comparison of the partial karyotypes of a t(5; 12) (q31q33; p12) associated with a del(5)(q31q33) and a microdeletion (12)(p12) found by FISH in pre-B ALL cell lines and a t(5; 12)(q33; p13) identified in cases with CMML. The arrows indicate the breakpoint regions on each derivative chromosome.
Comparison of the partial karyotypes of a t(5; 12) (q31q33; p12) associated with a del(5)(q31q33) and a microdeletion (12)(p12) found by FISH in pre-B ALL cell lines and a t(5; 12)(q33; p13) identified in cases with CMML. The arrows indicate the breakpoint regions on each derivative chromosome.
FISH was performed using several probes ordered along chromosomes 12p and 5q, and listed in Table 2. The chromosome 12p loci are ordered as follows: tel - D12S158 - 5′ETV6 - 3′ETV6 - D12S178 - D12S119 - D12S932 - D12S930 - KRAS2 - cen.12,13 The order of the chromosome 5 YACs is cen - y773D3 - y773B7 - y939F12 - y888A7 - y756A2 - y816D6 - y748D10 - y913F6 - tel.11 Probe y745D10 was also used for the present studies because it contains the PDGFRB gene and shows split signals on the der(5) and the der(12) when used for FISH analysis of the t(5; 12) in CMML.2 According to the CEPH YAC contig map14 y745D10 overlaps with y816D6, based on the presence of the RPS14 STS, and is located distally to y756A2. However, when the RPS14 STS was amplified from y745D10, we obtained a product with an aberrant size. According to the FISH findings (see below), the chromosome 5 sequences present in y745D10 are more likely to be located proximally to y756A2. Localization of IL3 (c33G8) and cC5I-20 to 5q31 and 5q34q35.1, respectively, was previously reported.15 16
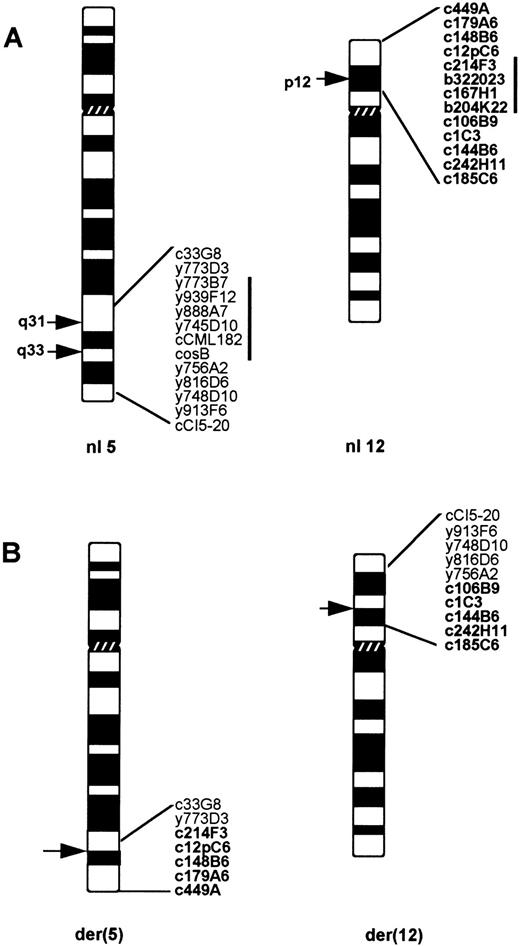
FISH results are summarized in Table 2 and schematically shown in Fig 2. The first experiments showed that in all three cell lines the breakpoint on 12p occurred between D12S178 and D12S119, clearly proximally to ETV6. To further refine this localization we isolated cosmid and BAC clones for four additional loci present in the interval flanked by D12S178 and D12S119,13 namely tel - D12S1094 - D12S1274 - D12S308 - GDI.D4 - cen. FISH analysis showed that the 12p breakpoint was flanked by GDI.D4 on the centromeric side and by D12S1094 on the telomeric side in the three cell lines. In addition, the sequences present in c167H1 or b204K22, both containing D12S308, and in b322023 (D12S1274) were lost from the der(12) and the der(5) in all cell lines. The consistent presence of fluorescent signals from all of these probes on the normal 12 excluded the possibility that the absence of signals on the derivative chromosomes was artifactual. The inserts of b204K22 and b322023, as measured by PFGE, are approximately 100 kb each. Thus, this is the lower size limit for the deleted region on 12p.
(A) The probes used for the FISH studies, ordered on the 5q31q35 and 12p12p13 regions of normal chromosomes 5 and 12. (B) Probes detected on each derivative chromosome of a t(5; 12)(q31q33; p12) in pre-B ALL. The probes derived from chromosome 12 are shown in bold. Note a deletion of 5q32 region flanked by y773D3 and y756A2, as well as a loss of 12p12 sequences between c12pC6 and c106B9.
(A) The probes used for the FISH studies, ordered on the 5q31q35 and 12p12p13 regions of normal chromosomes 5 and 12. (B) Probes detected on each derivative chromosome of a t(5; 12)(q31q33; p12) in pre-B ALL. The probes derived from chromosome 12 are shown in bold. Note a deletion of 5q32 region flanked by y773D3 and y756A2, as well as a loss of 12p12 sequences between c12pC6 and c106B9.
The analysis of the chromosome 5 breakpoint also yielded identical results for the three cell lines. As indicated in Table 2 and Fig 2, probes c33G8 and y773D3 containing IL3 or IL9, respectively, hybridized to the der(5) chromosome, whereas signals from y756A2, y816D6, y748D10, and y913F6 were detected on the der(12) in all cases. Unexpectedly, hybridization signals for y773B7, y939F12, y888A7, y745D10, as well as for cCML182 (CSF1R) and cosB (PDGFRB) probes, were present only on the normal chromosome 5 and absent on both the der(12) and the der(5) in all three cell lines. This excludes the involvement of PDGFRB in the eventual product of these translocations and shows that the translocations were accompanied by rearrangements of the chromosome 5 resulting in loss of material. Examples of FISH results are shown in Fig 3. Based on these findings, a description of a t(5; 12) in these cell lines should be corrected as follows: der(5)del(5)(q31q33)t(5; 12)(q31; p12), der(12)del(12)(p12p12)t(5; 12)(q33; p12).
Results of FISH analysis performed on the NALM-6 cell line with 5q ([A], y773D3; [C], cCML182; [E], y756A2) and 12p ([B],c106B9; [D], b204K22; [F], c214F3) derived probes. Note the presence of hybridization signals on the normal chromosomes 5 or 12 only with, respectively, cCML182 and b204K22. The signals of the centromeric chromosome 12 probe (pBR12, D12Z3) are shown in red, the signals of the cosmid, YAC, and BAC clones are shown in yellow. The arrowheads indicate the der(5) and the der(12).
Results of FISH analysis performed on the NALM-6 cell line with 5q ([A], y773D3; [C], cCML182; [E], y756A2) and 12p ([B],c106B9; [D], b204K22; [F], c214F3) derived probes. Note the presence of hybridization signals on the normal chromosomes 5 or 12 only with, respectively, cCML182 and b204K22. The signals of the centromeric chromosome 12 probe (pBR12, D12Z3) are shown in red, the signals of the cosmid, YAC, and BAC clones are shown in yellow. The arrowheads indicate the der(5) and the der(12).
Ohyashiki et al17 previously reported that the CSF1R locus was retained on the der(5) of the NALM-6 cell line. This was based on Southern experiments and in situ experiments with a radioactively labeled 1.4-kb CSF1R cDNA probe. However, the CSF1R probe used by these investigators detected EcoRI fragments of 6.5, 5.8, and 5.2 kbp upon Southern hybridization,17 whereas the genomic structure of the CSF1R gene predicts the detection of a 23-kbp fragment.18 To confirm this, we obtained the 1.4-kbp CSF1R cDNA probe and performed Southern with EcoRI-digested genomic DNA from the NALM-6, a control white blood cell sample and the EcoRI digested CSF1R cosmid. In all cases signals of a 23-kbp fragment were obtained.
DISCUSSION
Several recurrent chromosomal translocations were described for pre-B ALL, including t(1; 19)(q23; p13), t(4; 11)(q21; q23), t(9; 22)(q34; q11), and t(12; 21)(p13; q22), which are associated, respectively, with E2A-PBX, AF4-MLL, BCR-ABL, and ETV6-CBFA2 gene fusions.19,20 The latter has been detected in approximately 25% of pediatric patients with pre-B ALL and is now considered to be the most frequent abnormality in this subtype of leukemia in children.20 The occurrence of a t(5; 12), analogous to the one observed in a subgroup of CMML in four different pre-B ALL cell lines, triggered the question of whether an ETV6-PDGFRB fusion gene, previously shown to be involved in myeloid leukemia, might also play a role in the pathogenesis of lymphoblastic leukemia. To answer this question we performed an FISH analysis of the t(5; 12) present in the NALM-6, PBEI, and LR-10.6 pre-B ALL cell lines. Using a panel of 12p DNA probes, we show here that the 12p breakpoint of this translocation did not affect ETV6 but occurred in a more proximal region at p12, flanked by D12S1094 and GDI.D4. No expressed sequences are known within this segment of chromosome 12, and because many YAC clones assigned to this region apparently contain internal deletions, it is difficult to estimate the physical distance between the flanking markers. Moreover, in all three cell lines a cryptic microdeletion of D12S1274 and D12S308, mapped between D12S1094 and GDI.D4, was unexpectedly detected.
The situation on the der(5) also appears to be complex: the most distal probe hybridizing to the der(5) is y773D3 that contains the IL9 gene, which is at the telomeric end of the interleukin gene cluster but clearly proximal to PDGFRB, whereas all 5q33q35 probes including and distal to y756A2 are present on the der(12). Another intriguing observation is the absence of signals on either derivative chromosome with y745D10 and cosmid probes for PDGFRB or CSF1R (5q33), as well as with all YAC probes mapped between y773D3 and y756A2. These findings exclude the contribution of PDGFRB in a fusion event, as well as IL3 shown to be rearranged by a t(5; 14)(q31; q32) in pre-B ALL,21 and indicate that the translocation is also associated with the deletion of chromosome 5 sequences. The range of this deletion could not be measured; however, based on the YAC sizes it could span at least 5,350 kb. The FISH data were confirmed by reexamination of G-banded karyotypes of all three cell lines, which resulted in an identification of a previously overlooked deletion of a q32 band (see Figs 1 and 2).
The FISH data showing a loss of 5q31q33 sequences in analyzed cell lines are in conflict with the data previously reported on CSF1R for the NALM-6 cell line.17 As discussed above, the EcoRI fragments detected by these investigators on a genomic Southern blot are not in agreement with the known gene structure of CSF1R, suggesting that the cDNA probe used for these experiments was not the CSF1R cDNA fragment reported. This could then also be the explanation for the in situ results obtained by these investigators showing the presence of CSF1R on the der(5) of NALM-6 cells, whereas in our hands all analyzed sequences distal to those detected by y773D3 including CSF1R are absent from the der(5). It cannot be excluded that a secondary deletion occurred in the NALM-6 cell line during its maintenance in culture, but the consistent loss of the same region in the three cell lines analyzed here seems to argue against this possibility.
Deletion of the 5q31q33 and 12p12 sequences from both derivatives of a t(5; 12) is intriguing and the molecular consequences of these complex deletion/translocation events are unclear. It remains to be analyzed whether the deletions are independent from the translocation and affect gene(s) located within the breakpoint regions narrowed by FISH, or whether they are associated with the t(5; 12). In the latter case, the translocation might result in a rearrangement of two different loci that flank deletions on each chromosome and consequently produce two nonreciprocal chimeric fusions. A detailed physical mapping by the construction of cosmid and BAC contigs of the region is presently undertaken to answer these questions. Nevertheless, the systematic occurrence of these complex molecular rearrangements in all pre-B ALL cell lines studied here suggests that they could be directly related to the pathogenesis of this subtype of leukemia. The complex nature of these nonrandom anomalies then could explain why a t(5; 12) associated with a pre-B ALL is a very rare event.
Cryptic deletions of 12p have been previously detected in a wide range of hematologic malignancies including t(12; 21)-positive ALL.3 22-24 However, it should be noted that these are distinct from the deletion found here in pre-B ALL cells because almost all of them involve the region flanked by CDKN1B and ETV6 that remains untouched in the cell lines analyzed here.
On the other hand, a deletion of the long arm of chromosome 5 (5q−) is a frequent and well-known karyotypic abnormality reported in a broad spectrum of myeloid malignancies, including MDS and acute myeloid leukemia (AML).25,26 These cytogenetic findings led to the suggestion that a tumor suppressor gene, involved in normal myeloid growth and differentiation, is localized on the long arm of chromosome 5, more precisely at 5q31 that has been determined as the smallest region consistently lost in all patients with a del(5)(q).27 Although a few candidate tumor suppressor genes residing on 5q have been identified, no gene commonly inactivated in “5q−” cases has been found until now. Moreover, recent molecular studies suggest the presence of at least three distinct critical loci on 5q associated with the pathogenesis of myeloid leukemias. The first one, detected in preleukemic MDS and AML, is flanked by IL9 and EGR1 (size <2.4 mega base pair [Mbp])28,29; the second, typical for the “5q− syndrome” RA patients, is bordered by EGR1 and NKSF130; whereas the third one involves 5q13.1.31 Compilation of these and our data indicates that 5q31q33 deletion detected in NALM-6, PBEI, and LR-10.6 cell lines overlaps with one of the regions deleted in patients with myeloid disorders; however, whether the same genes are affected by del(5)(q) in myeloid and lymphoblastic leukemias remains unknown.
In summary, a t(5; 12)(q31q33; p12) in pre-B ALL does not affect the ETV6 or PDGFRB genes rearranged by an analogous translocation found in myeloid malignant disorders. The breakpoints of this translocation were mapped by FISH in the 5q31q33 and 12p12 regions, but the genes involved in the t(5; 12) have not been yet identified. Additionally, two cryptic deletions of 5q and 12p associated with the t(5; 12) have been found in all three pre-B ALL cell lines analyzed. These deletions, detected by FISH, are heterozygous and affect the chromosomes 5 and 12 that are involved in the t(5; 12) but not the other homologs. The 5q31q33 region of loss in the pre-B ALL cell lines overlaps with the deleted region observed in “5q-RA” patients, which seems to be distinct from the critical domains found in other MDS and AML patients.
ACKNOWLEDGMENT
We thank A. Hagemeijer (CME, Leuven, Belgium) and O. Bernard (INSERM U409, Paris, France) for providing us with NALM-6 and PBEI cell lines, M. Lovett (University of Texas Southwestern Medical Center at Dallas) for the IL3 (c33G8) probe, M. Roberts (M.D. Anderson Cancer Center, The University of Texas) for the cosB probe, A. Roebroek (Center for Human Genetics, Leuven, Belgium) for the cCML182 and CSF1R cDNA probes, Y. Nakamura (Cancer Institute, Tokyo, Japan) for the cC5-20 probe, and M. Dehaen for cytogenetic analysis and help in the preparation of this manuscript.
Supported in part by the Grants No. VIS 94/16 and G.0153.96 of the Foonds vor Wetenschappelijk Onderzoek (F.W.O.). P.M. is an 'onderzoeksdirecteur' of the Nationaal Fonds voor Wetenschappelijk Onderzoek, Belgium.
Address reprint requests to Peter Marynen, PhD, Center for Human Genetics, University of Leuven, Campus Gasthuisberg O&N6, Herestraat 49, B-3000 Leuven, Belgium.

![Fig. 3. Results of FISH analysis performed on the NALM-6 cell line with 5q ([A], y773D3; [C], cCML182; [E], y756A2) and 12p ([B],c106B9; [D], b204K22; [F], c214F3) derived probes. Note the presence of hybridization signals on the normal chromosomes 5 or 12 only with, respectively, cCML182 and b204K22. The signals of the centromeric chromosome 12 probe (pBR12, D12Z3) are shown in red, the signals of the cosmid, YAC, and BAC clones are shown in yellow. The arrowheads indicate the der(5) and the der(12).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/5/10.1182_blood.v89.5.1716/3/m_bl_0043f3.jpeg?Expires=1769176519&Signature=FcaDW-lp-fCTJh-ul5orxgMYlQogBvdSypV1F8~2cRahHA7dhceJxEyam~spPZPtI-Fww~Vi~nnebCdOs4PZR36Qgyc~7QKxjA5yjIQGbRV~bCulNSOgTFW-jPSR2kHKwQS6kFavg50uKjR3uGGqT80QMFEwHFQKHooSbpF~ozUVbAp78-JB0uPEPSiCglpDqmL4jj2xfoMisXfyM~eZp4f-iphpUj8FcL1agvAlpOBOKqunDbIqZTpvv6qGWXzgQKT-wDEz8AB67uoYwNUwgpiV07NIEgQjdBseXbMA~oSmieTKLDitFD-Sk8aErwNKZQKMwNkHxv1VWW1z3x3viQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal