Abstract
The hemophilia A mutation database lists more than 160 missense mutations: each represents a molecular defect in the FVIII molecule, resulting in the X-linked bleeding disorder hemophilia A with a clinical presentation varying from mild to severe. Without a three-dimensional FVIII structure it is in most cases impossible to explain biological dysfunction in terms of the underlying molecular pathology. However, recently the crystal structure of the homologous human plasma copper-binding protein ceruloplasmin (hCp) has been solved, and the A domains of FVIII share approximately 34% sequence identity with hCp. This advance has enabled the building of a molecular model of the A domains of FVIII based on the sequence identity between the two proteins. The model allows exploration of predictions regarding the general features of the FVIII molecule, such as the binding-sites for factor IXa and activated protein C; it has also allowed the mapping of more than 30 selected mutations with known phenotype from the database, and the prediction of hypothetical links to dysfunction in all but a few cases. A computer-generated molecular model such as that reported here cannot substitute for a crystal structure. However, until such a structure for FVIII becomes available, the model represents a significant advance in modeling FVIII; it should prove a useful tool for exploiting the increasing amount of information in the hemophilia A mutation database, and for selecting appropriate targets for investigation of the structure-function relationships via mutagenesis and expression in vitro.
FACTOR VIII (FVIII) plays an essential role as a cofactor in blood coagulation1: absence of functional FVIII is responsible for the commonest bleeding disorder, hemophilia A. The extensive database of FVIII mutations correlating with clinically-defined bleeding underlines the central regulatory role of FVIII in the hemostatic network.2 FVIII is synthesized in hepatocytes as a mature single-chain polypeptide of 2,332 amino acids (predicted Mr = 264,763), but circulates in plasma as a population of noncovalently linked heterodimers in a procofactor form bound to von Willebrand factor (vWF ). During coagulation, FVIII is proteolytically activated to FVIIIa by trace amounts of thrombin or factor Xa (FXa) and greatly accelerates the proteolysis of factor X to FXa by factor IXa (FIXa) on the activated platelet surface. In complex with another phospholipid-bound cofactor, factor Va (FVa), FXa cleaves prothrombin to thrombin; FVa is homologous to FVIII in both structure and function. Finally, thrombin cleaves fibrinogen to insoluble fibrin to stabilize a hemostatic clot. Feedback control is mediated by activated protein C (APC), which inactivates FVIIIa and FVa by further proteolysis.
FVIII is a large multidomain protein containing internal repeats.3 There are three homologous A-type domains (A1, A2, and A3) defined approximately by residue positions 1-336, 375-719, and 1691-2025, respectively. An acidic peptide a1 spans 337-374 and separates A1 from A2. A second acidic peptide, a2, (720-740) connects A2 with the large, heavily-glycosylated B domain, which encompasses approximately residues 741-1648. This leads to a third short acidic peptide, here termed a3, (1649-1690), which itself connects with the A3 domain. Finally, there are two homologous C-terminal domains (C1 and C2) each of approximately 155 amino acids. The domain organization of the FVIII polypeptide can thus be designated as A1.a1.A2.a2.B.a3.A3.C1.C2. The location of seven disulphide bonds within the FVIII molecule has been reported: there are two in each of the A1, A2, and A3 domains and one within the C1 domain.4
On or before secretion proteolytic processing occurs within the B domain to give heterodimers consisting of a 90- to 220-kD heavy chain A1.a1.A2.a2 with variable extensions of the B domain, associated via a divalent cation-dependent interaction with an 80-kD light chain consisting of a3.A3.C1.C2.5 During coagulation thrombin cleaves the heterodimers at three points to form FVIIIa: after R372 near the N-terminus of A2, R740 at the N-terminus of B, and R1689 at the N-terminus of A3.6 Critical for FVIIIa activity, fragment A2.a2 remains noncovalently associated with the A1.a1 and A3.C1.C2 polypeptides, while the variable B fragments and the short a3 peptide are lost; thus, FVIIIa is a heterotrimer that may be designated A1.a1*A2.a2 *A3.C1.C2.
FVIII may also be cleaved by FXa after residues R336, R372, R740, R1689, and R1721,6 and by FIXa after residues R336 and R1719.7 All these specific cleavages either promote or degrade the functional activity of FVIII/FVIIIa, thus, its activity in coagulation is closely regulated by limited proteolysis. APC mediates negative feedback destruction of FVIIIa activity by cleavage after R336 and R562.8
Various interaction sites have been defined on FVIII. Two FIXa binding-sites have been localized to the vicinity of residues S558-Q565 in A2 and E1811-K1818 in A3.9,10 Recent work has shown that the A3 site binds to the FIXa EGF1 domain,11 whereas the A2 site binds to the serine protease domain.12 In addition, an APC binding-site has been described in A3 within H2007-V2016.13 FVIII(a) binds to phospholipid surfaces via sequence(s) in the C2 domain, and to its carrier vWF via the a3 peptide, and possibly also C2.14,15 A recent report also identified a region of the C-terminal portion of A1 (T291-F309), which may be involved in binding to the chaperonin BiP; this protein is associated with inhibition of FVIII secretion.16
Amino acid sequence lineups of human FVIII with other A domain proteins. (A through C) Alignment of human FVIII A1 (A), A2 (B), and A3 (C) domains with hCp, showing SCR and VR placing together with conserved residues. Lowercase, Vrs; uppercase with bar above sequence, SCRs. Sequence numberings refer to the mature proteins. (D through F ) Alignment of A1 (D), A2 (E), and A3 (F ) domain sequences of: human and mouse FVIII (humf8,murf8); human and bovine FV (humfv,bovfv); rat and human Cp (ratcp,humcp), showing extent of conservation of human FVIII residues with these related proteins. Key: white characters on black background, identical in all six sequences; white on grey, residues identical to that in human FVIII; black on grey, residue similar to that in human FVIII; black on white, residues dissimilar to human FVIII.
Amino acid sequence lineups of human FVIII with other A domain proteins. (A through C) Alignment of human FVIII A1 (A), A2 (B), and A3 (C) domains with hCp, showing SCR and VR placing together with conserved residues. Lowercase, Vrs; uppercase with bar above sequence, SCRs. Sequence numberings refer to the mature proteins. (D through F ) Alignment of A1 (D), A2 (E), and A3 (F ) domain sequences of: human and mouse FVIII (humf8,murf8); human and bovine FV (humfv,bovfv); rat and human Cp (ratcp,humcp), showing extent of conservation of human FVIII residues with these related proteins. Key: white characters on black background, identical in all six sequences; white on grey, residues identical to that in human FVIII; black on grey, residue similar to that in human FVIII; black on white, residues dissimilar to human FVIII.
FVIII shows clear homology to FV; the latter has a domain structure A1.A2.B.A3.C1.C2. There is approximately 35% to 40% sequence identity between all the corresponding domains of FVIII and FV, with the exception of the B domains, which are apparently unrelated.17 Additionally, FV lacks the short acidic sequences a1, a2, and a3 found in FVIII. The blue copper-binding plasma protein ceruloplasmin (Cp) is also homologous to FVIII and FV.3,18 The three A domains of Cp show approximately 34% sequence identity with the FVIII A domains. A crystal structure of human Cp (hCp) has been solved, showing that the A domains are arranged in a triangular structure, with each domain made up of two cupredoxin-type folds.19 We have used the coordinates of the hCp crystal structure to build a model of the A domains of FVIII, using homology modeling techniques. The third edition of the hemophilia A mutation database is now accessible as a World Wide Web site2 (URL: http://europium. mrc.rpms.ac.uk) and has been used here with the model to investigate hemophilic mutations; it may also be used to direct experimental studies, which will shed light on the molecular pathology of hemophilia A. In addition, modeling of the structure with FIXa binding-sites leads to a hypothesis for the general configuration of the factor X-activating complex.
MATERIALS AND METHODS
A model of the A1, A2, and A3 domains of FVIII was built using the Homology module (version 2.3) of the Insight II modeling suite (Biosym/MSI, San Diego, CA). The X-ray coordinates of hCp were imported and displayed with its associated linear amino-acid sequence. The sequences of the A domains of FVIII were imported as a single polypeptide, and aligned with the hCp sequence as previously reported.3
The set of primary-structure conserved regions (SCRs) were defined using this alignment (Fig 1, A through C), and the coordinates of the hCp crystal structure were then imposed on the FVIII sequence. Insertions and deletions were contained within variable regions (VRs). A search of the Brookhaven Protein Database generated a selection of peptide loops in which the distance between the N- and C-termini was similar to that in each of the VRs. These candidate loops varied in the amount of overlap between their N-and C-termini and those of the corresponding VR, thus each potential loop was scrutinized for the best fit within the model. Loops with the closest overlap which also lay on the surface of the FVIII model were chosen.
The coordinates of these loops were then superimposed to give an α-carbon backbone for each VR, and the sidechains of each residue were then varied to correspond with the FVIII sequence within the VR. Finally, coordinates from the Insight II amino acid library were imposed on the short N- and C-termini which lay beyond the first and last SCR, respectively.
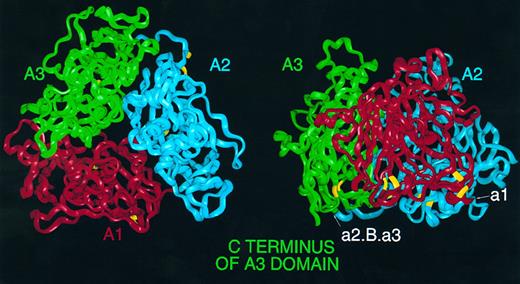
Homology model of the triplicated A domains of human FVIII. (Left) FVIII α-carbon ribbon viewed from 'top' of the molecule down the pseudo threefold axis. A1 (red), A2 (blue), and A3 (green); this color coding is consistent throughout this and subsequent figures. Cysteine residues predicted to participate in disulphide links are highlighted in yellow. (Right) View perpendicular to the axis, with A1 to the front and the 'top' of the molecule bearing the loops anchoring the first and second β-strands of subdomains corresponding to the d subdomains of hCp; the A3 C terminus is marked to the left. Also marked are the location of insertion points of acidic peptides in FVIII; peptide a1 may be inserted on the surface between A1 and A2, whereas peptide a2, B domain and third acidic peptide a3 may be inserted on surface between A2 and A3.
Homology model of the triplicated A domains of human FVIII. (Left) FVIII α-carbon ribbon viewed from 'top' of the molecule down the pseudo threefold axis. A1 (red), A2 (blue), and A3 (green); this color coding is consistent throughout this and subsequent figures. Cysteine residues predicted to participate in disulphide links are highlighted in yellow. (Right) View perpendicular to the axis, with A1 to the front and the 'top' of the molecule bearing the loops anchoring the first and second β-strands of subdomains corresponding to the d subdomains of hCp; the A3 C terminus is marked to the left. Also marked are the location of insertion points of acidic peptides in FVIII; peptide a1 may be inserted on the surface between A1 and A2, whereas peptide a2, B domain and third acidic peptide a3 may be inserted on surface between A2 and A3.
Each domain was then examined for serious steric overlaps. These were relieved as far as possible by selecting lower-energy conformations from a library of rotamers within the program. The molecule was then energy-minimized in stages. The splice points between each VR and SCR were the most strained regions of the model, so these were minimized first as a group using the standard steepest descents algorithm to give an acceptably low energy derivative of around 10 kcal.mol−1. The rest of the molecule was then minimized in the order: N- and C-termini, VRs, and finally SCRs.
Finally the molecule was energy-minimized, one domain at a time, with the Discover module (Biosym/MSI, version 3.1) of Insight II, using the steepest descents algorithm until a low energy derivative of around 1 kcal.mole−1 had been reached; at this point the calculated total energy of the molecule was consistent with a relaxed and stable conformation.
To examine the possible effects on the model of selected missense mutations reported in the hemophilia A database, residue sidechains were altered using the Biopolymer module of Insight II and, where appropriate, the variant molecules were reminimized using Discover.
RESULTS AND DISCUSSION
A homology-based molecular model of the A domains of FVIII based on the crystal structure of the copper-binding protein nitrite reductase (NIR) has recently been proposed.26 NIR, which is much more distantly related to FVIII than is Cp, crystalizes as a triangular homotrimer whose three identical A domains each consist of a two-subdomain d1.d2 structure (whose d-type subdomain sequences are themselves related). Although useful in terms of prediction of the hypothetical general shape of FVIII, this previous model possesses two serious defects. Firstly, the low level of identity between NIR and FVIII A domains (varying between only 11% and a maximum of 28%) renders modeling of other than broad features very suspect; secondly, the arrangement of subdomains is almost certainly incorrect, with the C-terminal d subdomains of each FVIII A domain pointing outwards toward the top of the molecule rather than inwards as predicted by comparison with Cp,19 because of the alignment of these subdomains' sequences with the N-terminal d1 subunit of NIR rather than the C-terminal d2 subunit.
General arrangement.The crystal structure of hCp consists of three A domains in a triangular array showing a clear pseudo-threefold axis.19 Each A domain is formed from two related d subdomains with d1 at the N-terminus of hCp and d6 at the C-terminus: each d subdomain resembles the fold of a typical cupredoxin-like molecule and consists of a β-barrel structure.27 The first and second β-strands of each subdomain are linked by a large loop lying at the ‘top’ of the domain, with the loop conformations appearing very different for the odd and even d subdomains. The odd-numbered subdomains d1, d3, and d5 all point outwards towards the ‘top’ of the pseudo-threefold axis, whereas the even-numbered subdomains d2, d4 and d6, which contain the mononuclear type-I copper atoms, point inwards.19 In NIR the mononuclear type-I copper atoms are sited in the N-terminal d1 domains so that they are near the outside of the molecule. This is consistent with NIR participating in electron transfer with pseudoazurin, a reasonably substantial molecule of some 130 amino acids. However, the location of the copper atoms in the even d subdomain of hCP (and therefore towards the inside of the molecule) implies that interactions with hCP are mediated by small molecules.
The overall structure of the FVIII A domain model we have constructed by homology with hCp also shows these broad features (Figs 2 and 3): the A1, A2, and A3 domains correspond to d1.d2, d3.d4, and d5.d6 in hCp and the ‘top’ of the molecule is covered by the loops which connect the first two β-strands in each of the six d subdomains. The d2, d4, and d6 subdomains point inwards toward the top of the molecule, unlike in the NIR-based model of FVIII where these subdomains point outwards.26
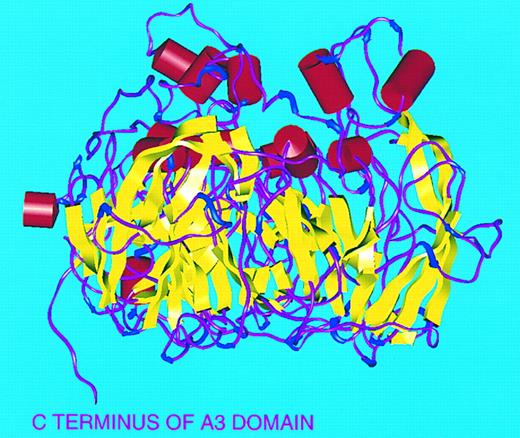
Secondary structure of FVIII A domains. Viewed perpendicular to threefold axis with ‘top’ of molecule to top of figure. α-Helices (red), β-sheets (yellow), turns (blue), and random coil (magenta).
Secondary structure of FVIII A domains. Viewed perpendicular to threefold axis with ‘top’ of molecule to top of figure. α-Helices (red), β-sheets (yellow), turns (blue), and random coil (magenta).
Disulphide links.Of the six disulphide links identified in the FVIII A domains,4 five (A1: 153-179, 248-329; A2: 528-554, 630-711; A3: 1832-1858) are homologous to those in hCp and can be placed at the ‘bottom’ of the model. The sixth A domain disulphide (A3: 1899-1903) is nonhomologous and is predicted to be further towards the top of the model.
Proteolytic cleavage sites.The FVIII acidic peptides a1, a2, a3, and the large B domain all contain function-related cleavage sites at one or both of their N- or C-termini, and thus these sites would be predicted to be accessible to serine proteases such as thrombin and APC. Although these peptide sequences are far too large to be modeled realistically, the FVIII A domain model can predict the location of the termini of these loops, which must be surface-exposed. Thus, the FVIII model predicts that the N- and C-termini of the first acidic region a1 (330-379) lie on the surface of the molecule at the interface between A1 and A2 (Fig 2). Similarly, the interface between A2 and A3 (into which the second acidic region a2, the B domain, and the third acidic region a3 must be inserted) also lies on the surface (Fig 2). Predictions such as these and others described below help to support the functional significance of the model.
The copper atom in FVIII.Six integral copper atoms have been positively identified in the hCp crystal structure.19 Three are type-I blue copper atoms, found in homologous positions in subdomains d2, d4, and d6. The remaining three copper atoms are located in a trinuclear cluster at the interface between subdomains d1 and d6, that is, between A1 and A3: two of these are type-III copper atoms, each coordinated to three histidine residues (to H103 and H161 in A1, and H1022 in A3; to H163 in A1, and to H980 and H1020 in A3). The third atom in the cluster is type-II and is coordinated to H101 in A1 and H978 in A3. On close examination of the electron density of the trinuclear cluster in hCp, a similarity is seen to the layout of the homologous trinuclear centre in the crystal structure of ascorbate oxidase (AO); in AO, one oxygen heteroatom bridges the type-III copper atoms and a second coordinates to the type-II copper atom.28 Thus (based on analogy with AO) it is likely that in the crystal structure of hCp, this latter oxygen heteroatom can form hydrogen bonds with the sidechain hydroxyl group of Y107 and the main chain carbonyl oxygen of S102.
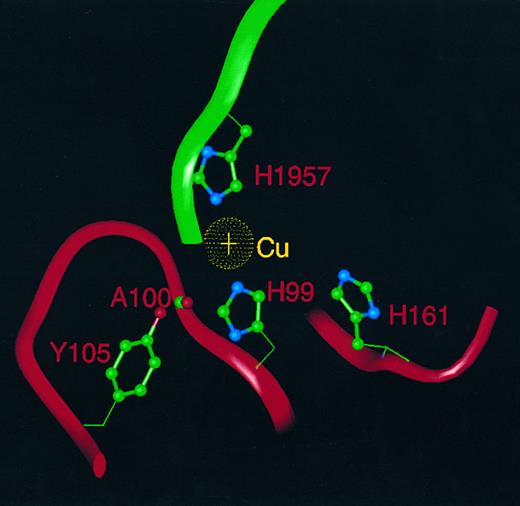
FVIII has been shown to contain one copper atom per molecule,29 and therefore it is likely that the position of this copper atom in the molecule will correspond to one of the six copper atoms identified in the crystal structure of hCp. Because purified FVIII protein shows none of the spectral characteristics typical of a type-I copper-binding protein, this rules out occupancy of either of the conserved type-I sites in subdomains d2 and d6. Of the six histidine ligands that coordinate the two type-III coppers in the trinuclear cluster of hCp, only one (H163 in hCp) is conserved in FVIII as H161. However, the two histidine residues (H101, H978) that coordinate to the type-II copper in the hCp trinuclear cluster are conserved as H99 and H1957 in FVIII, as is the aforementioned tyrosine (Y107) residue as Y106 in FVIII. We suggest that the single copper atom in FVIII lies at the interface between domains A1 and A3 (between subdomains d1 and d6) homologous to the position of the type-II copper in hCp, coordinated by H99 in A1 and H1957 in A3. H161 in A1 (postulated in the earlier study26 to be a third histidine ligand), is too far displaced from the others to act as a copper ligand in our model. It is possible that the third ligand may (by analogy with the hCp structure) be an oxygen atom of a water molecule, which is hydrogen-bonded both to the sidechain hydroxyl of Y106 and to the main chain carbonyl oxygen of A100 (Fig 4). The interdomain contact mediated by the copper atom may contribute to the association of the heavy and light chains of FVIII through a divalent cation-dependent interaction involving A1 and A3.
The copper atom in FVIII and its ligands. A single copper atom (gold) is shown with ligands H99 and H1957, whereas an oxygen heteroatom (not shown) of water or hydroxide acts as a third ligand and is hydrogen bonded to the A100 main chain carbonyl and the sidechain hydroxyl of Y105. H161 is too distant to act as a copper ligand. Neighboring residues are displayed as α-carbon ribbons: A1 (red) and A3 (green).
The copper atom in FVIII and its ligands. A single copper atom (gold) is shown with ligands H99 and H1957, whereas an oxygen heteroatom (not shown) of water or hydroxide acts as a third ligand and is hydrogen bonded to the A100 main chain carbonyl and the sidechain hydroxyl of Y105. H161 is too distant to act as a copper ligand. Neighboring residues are displayed as α-carbon ribbons: A1 (red) and A3 (green).
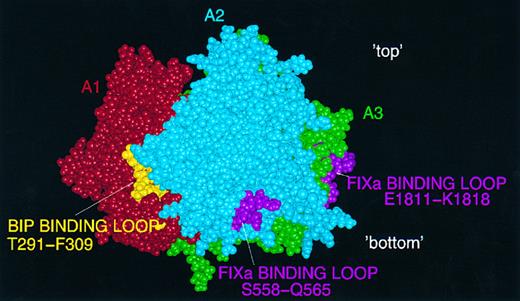
Protein interaction sites.Amino acid residues implicated in the two binding sites identified recently for FIXa9,10 lie on the surface of the model, as would be expected (Fig 5). Also shown in this figure is the putative BiP binding-site within T291-F309,16 which appears as a partly exposed region rich in hydrophobic residues. The sequence within which the suggested binding site for APC lies is toward the C-terminus of the A3 domain, and is partly buried in the model (Fig 6).13 Finally, two tyrosine residues in the model which are known to be posttranslationally sulphated30 (Y718 and Y719) lie close to the surface at the interface between A2 and A3 (Fig 6). These residues may play a role in thrombin binding via its anion-binding exosite, which is known to interact strongly with sulphated tyrosine (eg, in hirudin). It should be noted that our model of the FVIII A2 domain terminates at Y719 and that the predicted structure here should be regarded with particular caution.
Predicted binding-sites for FIXa and BiP on the FVIII A domain model. FIXa: two loops, in A2 (S558-Q565) and A3 (E1811-K1818) are shown in pink on the surface of a CPK FVIII image: view perpendicular to the threefold axis with 'top' and 'bottom' of the molecule marked. Also shown is the partly exposed putative BiP binding site (T291-F309) in gold.
Predicted binding-sites for FIXa and BiP on the FVIII A domain model. FIXa: two loops, in A2 (S558-Q565) and A3 (E1811-K1818) are shown in pink on the surface of a CPK FVIII image: view perpendicular to the threefold axis with 'top' and 'bottom' of the molecule marked. Also shown is the partly exposed putative BiP binding site (T291-F309) in gold.
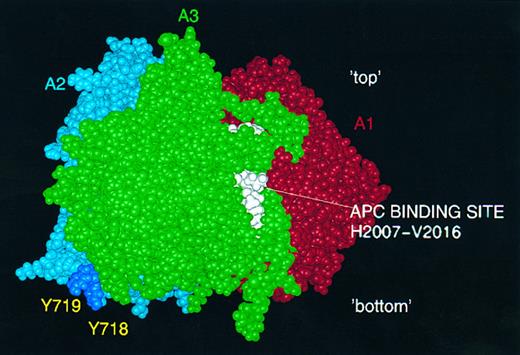
Predicted activated protein C (APC) and thrombin interaction sites. APC binding loop (H2007-V2016) on the A3 surface is shown in white: postulated thrombin interaction via sulphated tyrosine residues Y718 and Y719 (deep blue) in FVIII A2 domain.
Predicted activated protein C (APC) and thrombin interaction sites. APC binding loop (H2007-V2016) on the A3 surface is shown in white: postulated thrombin interaction via sulphated tyrosine residues Y718 and Y719 (deep blue) in FVIII A2 domain.
Interpretation of mutations implicated in clinical disease.The possible effects on the FVIII A domain model of 36 single missense mutations from the hemophilia A database have been examined (Table 1). These were only those mutations for which circulating FVIII antigen (FVIII:Ag) assay levels are reported and, in the absence of three-dimensional structural information, they may be conveniently divided into two main classes. Firstly, cross-reacting material positive (CRM+): circulating FVIII:Ag levels normal (>50%) with biological activity (FVIII:C) reduced (<50%). These are dysfunctional variants - the mutation may affect a functional region but still allows the molecule to fold, be secreted, and circulate in a stable form. Study of these dysfunctional mutants allows mapping of functional regions on the model, and experimentally verifiable predictions can be made. Secondly, cross-reacting material negative (CRM−): both FVIII:Ag and FVIII:C levels are sharply reduced, indicating that these mutations are severe enough to interfere with the secretion of the molecule or its circulation in a stable form; such molecules may or may not be functionally deficient in addition.
Table of 36 Missense Mutations Studied, With FVIII:C and FVIII:Ag Levels, Clinical Severity, Level of Conservation and Comments on Possible Molecular Pathology
| Mutation . | ID* . | FVIII:C (%) . | FVIII:Ag (%) . | Clinical Severity . | Conservation† . | Hypothetical Pathology . |
|---|---|---|---|---|---|---|
| Y114C | HP19 | 6.3 | 10.7 | Mild | YYYYYY | Free Cys hinders correct folding |
| T1181 | HP20 | 2.0 | 10.7 | Moderate | TTTTTT | Steric clash with D116 in core |
| V162M | HP25 | 5.3 | 14 | Mild | VVEVVI | No assignable reason for phenotype |
| K166T | JH139 | 8.0 | 9.8 | Mild | KKEEKK | Loss of positive charge |
| S170L | LKC | 3.5 | 8.7 | Mild | SSSSSS | Strain in core, possible loss of H-bond with K166 main chain carbonyl |
| D203V | HP28 | 2.0 | 8.5 | Mild | DDDDDD | Loss of negative charge |
| E272G | JH20 | 2.0 | 3.5 | Moderate | EENNHH | Possible loss of H-bond with L307 main chain carbonyl |
| T275I | HP29 | 4.8 | 20.2 | Moderate | TTVVAA | Possible loss H-bond with H274 main chain carbonyl |
| R282H | JH86 | <1 | 18 | Severe | RRKKHR | Modification of A1/A2 interaction via D525 |
| S289L | JH152 | 37 | 106 | Mild | SSVVFF | Modification of A1/A3 interaction via Y1979 |
| T295A | HP32 | 9.5 | 11.6 | Mild | TTTTDD | No assignable reason for phenotype |
| C329S | Lisboa2 | 2.6 | 3.2 | Moderate | CCCCCC | Loss of conserved disulphide link |
| L412F | JH131 | 7.0 | 6.4 | Mild | LLLLFF | Steric clash with S409 in core |
| K425R | JH74 | <1 | 5.0 | Severe | KKKKKK | Steric strain in core |
| A469G | HP38 | 2.3 | 45.3 | Moderate | AAAAGG | No assignable reason for phenotype |
| I475T | HP39 | 5.5 | 8.8 | Mild | IIIIII | No assignable reason for phenotype |
| G479R | Porto1 | 17.8 | 31.6 | Mild | GGGGGG | Steric clash with R531 in core |
| D525N | MS | 6.0 | 61 | Moderate | DDDDDD | Modification of A1/A2 interaction |
| R527W | DG | 15.0 | 126 | Mild | RRQQVV | Disturbance of FIXa binding-site |
| R531H | HP46 | 23.5 | 33.2 | Mild | RRRRKK | Modification of A1/A2 interaction via R282 |
| D542G | JH63 | <1 | 5.0 | Severe | DDDDDD | Loss of negative charge |
| S558F | JH151 | 21 | 175 | Mild | SSSSSS | Steric clash with C554/Y555 round FIXa binding-site |
| I566T | FW | 4 | 200 | Moderate | IMRRKK | Predicted new N-glycosylation at N564 close to FIXa binding-site |
| V634A | JH156 | 5 | 138 | Mild | VVTTSS | No assignable reason for phenotype |
| A644V | JH136 | 14 | 25 | Mild | AATTNN | Modification of A1/A2 interaction |
| F658L | HP49 | 5.1 | 50.5 | Mild | FFFFYY | Decrease in core volume |
| A704T | HP51 | 4.5 | 5.5 | Moderate | AALLQQ | No assignable reason for phenotype |
| G1750R | HP77 | 22 | 23.5 | Mild | GGGGRK | Addition of positive charge and bulky sidechain |
| L1756V | HP83 | 5.0 | 1.5 | Mild | LLLLLL | No assignable reason for phenotype |
| M1772T | JH116 | <1 | 72 | Severe | MMQQKK | Predicted new N-glycosylation at N1770 close to FIXa binding-site |
| R1781H | HP84 | 2.0 | 4.7 | Moderate | RRRRRR | No assignable reason for phenotype |
| P1825S | HP88 | 12 | 18.1 | Mild | PPPPAA | Close to FIXa binding-site |
| H1848R | HP89 | 1.5 | 20.1 | Moderate | HHHHIY | Modification of A2/A3 interaction via D696 |
| R1941Q | JH33 | 4.5 | 20 | Mild | RRRRNN | Close to FIXa binding-site |
| G1948D | Porto2 | 7.4 | 48.7 | Mild | GGGGGG | Close to A2/A3 interface |
| H1961Y | HP91 | 10.5 | 7.8 | Mild | HHQQHH | Increases core energy |
| Mutation . | ID* . | FVIII:C (%) . | FVIII:Ag (%) . | Clinical Severity . | Conservation† . | Hypothetical Pathology . |
|---|---|---|---|---|---|---|
| Y114C | HP19 | 6.3 | 10.7 | Mild | YYYYYY | Free Cys hinders correct folding |
| T1181 | HP20 | 2.0 | 10.7 | Moderate | TTTTTT | Steric clash with D116 in core |
| V162M | HP25 | 5.3 | 14 | Mild | VVEVVI | No assignable reason for phenotype |
| K166T | JH139 | 8.0 | 9.8 | Mild | KKEEKK | Loss of positive charge |
| S170L | LKC | 3.5 | 8.7 | Mild | SSSSSS | Strain in core, possible loss of H-bond with K166 main chain carbonyl |
| D203V | HP28 | 2.0 | 8.5 | Mild | DDDDDD | Loss of negative charge |
| E272G | JH20 | 2.0 | 3.5 | Moderate | EENNHH | Possible loss of H-bond with L307 main chain carbonyl |
| T275I | HP29 | 4.8 | 20.2 | Moderate | TTVVAA | Possible loss H-bond with H274 main chain carbonyl |
| R282H | JH86 | <1 | 18 | Severe | RRKKHR | Modification of A1/A2 interaction via D525 |
| S289L | JH152 | 37 | 106 | Mild | SSVVFF | Modification of A1/A3 interaction via Y1979 |
| T295A | HP32 | 9.5 | 11.6 | Mild | TTTTDD | No assignable reason for phenotype |
| C329S | Lisboa2 | 2.6 | 3.2 | Moderate | CCCCCC | Loss of conserved disulphide link |
| L412F | JH131 | 7.0 | 6.4 | Mild | LLLLFF | Steric clash with S409 in core |
| K425R | JH74 | <1 | 5.0 | Severe | KKKKKK | Steric strain in core |
| A469G | HP38 | 2.3 | 45.3 | Moderate | AAAAGG | No assignable reason for phenotype |
| I475T | HP39 | 5.5 | 8.8 | Mild | IIIIII | No assignable reason for phenotype |
| G479R | Porto1 | 17.8 | 31.6 | Mild | GGGGGG | Steric clash with R531 in core |
| D525N | MS | 6.0 | 61 | Moderate | DDDDDD | Modification of A1/A2 interaction |
| R527W | DG | 15.0 | 126 | Mild | RRQQVV | Disturbance of FIXa binding-site |
| R531H | HP46 | 23.5 | 33.2 | Mild | RRRRKK | Modification of A1/A2 interaction via R282 |
| D542G | JH63 | <1 | 5.0 | Severe | DDDDDD | Loss of negative charge |
| S558F | JH151 | 21 | 175 | Mild | SSSSSS | Steric clash with C554/Y555 round FIXa binding-site |
| I566T | FW | 4 | 200 | Moderate | IMRRKK | Predicted new N-glycosylation at N564 close to FIXa binding-site |
| V634A | JH156 | 5 | 138 | Mild | VVTTSS | No assignable reason for phenotype |
| A644V | JH136 | 14 | 25 | Mild | AATTNN | Modification of A1/A2 interaction |
| F658L | HP49 | 5.1 | 50.5 | Mild | FFFFYY | Decrease in core volume |
| A704T | HP51 | 4.5 | 5.5 | Moderate | AALLQQ | No assignable reason for phenotype |
| G1750R | HP77 | 22 | 23.5 | Mild | GGGGRK | Addition of positive charge and bulky sidechain |
| L1756V | HP83 | 5.0 | 1.5 | Mild | LLLLLL | No assignable reason for phenotype |
| M1772T | JH116 | <1 | 72 | Severe | MMQQKK | Predicted new N-glycosylation at N1770 close to FIXa binding-site |
| R1781H | HP84 | 2.0 | 4.7 | Moderate | RRRRRR | No assignable reason for phenotype |
| P1825S | HP88 | 12 | 18.1 | Mild | PPPPAA | Close to FIXa binding-site |
| H1848R | HP89 | 1.5 | 20.1 | Moderate | HHHHIY | Modification of A2/A3 interaction via D696 |
| R1941Q | JH33 | 4.5 | 20 | Mild | RRRRNN | Close to FIXa binding-site |
| G1948D | Porto2 | 7.4 | 48.7 | Mild | GGGGGG | Close to A2/A3 interface |
| H1961Y | HP91 | 10.5 | 7.8 | Mild | HHQQHH | Increases core energy |
Unique patient identifier: where multiple reports occur in the hemophilia A database, only the first entry with full phenotype is given here.
This column gives the single letter amino-acid code for the wild-type residues in this position in (L-R): human FVIII, murine FVIII, human FV, bovine FV, rat Cp, and human Cp.
Three mutations with plausible mechanisms for clinical disease based on the FVIII A domain model are briefly discussed below: to allow inspection of the level of conservation of all A domain residues in human FVIII with murine FVIII, human and bovine FV, and human and rat Cp, multiple sequence lineups for the three A domains are presented in Fig 1 (D through F ).
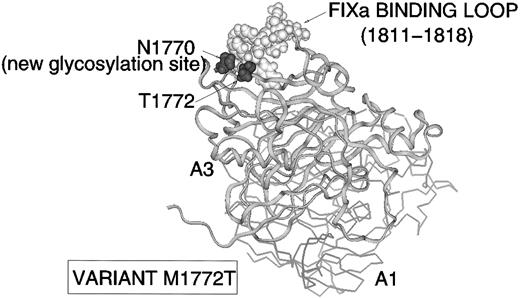
M1772T (FVIII:C <1 %, FVIII:Ag 72%; CRM+) is an example of a mutation affecting a predicted FIXa interaction site. Replacement of M1772 by Thr creates a new glycosylation consensus sequence (N-X-S/T): the N1770 sidechain is surface-exposed (Fig 7) and thus potentially available for glycosylation. The model indicates that N1770 lies close to the proposed binding-site for FIXa within E1811-K181810: N1770 is a FVIII-specific residue further implicating this region in FVIII cofactor function.
Variant M1772T. A3 α-carbon backbone ribbon (and line trace of A1) showing location of A3 FIXa binding site (E1811-K1818, white CPK) with new glycosylation site N1770 (black CPK) created in the vicinity by mutation of M1772 (black CPK) to Thr.
Variant M1772T. A3 α-carbon backbone ribbon (and line trace of A1) showing location of A3 FIXa binding site (E1811-K1818, white CPK) with new glycosylation site N1770 (black CPK) created in the vicinity by mutation of M1772 (black CPK) to Thr.
G1948D (FVIII:C 7.4%, FVIII:Ag 48.7%; CRM+) is a mutation postulated to affect interdomain interaction. G1948 lies in the core of the molecule near the interface of the A2 and A3 domains and on a turn between two β-strands, and is an absolutely conserved residue. Replacement with the bulky Asp sidechain introduces significant steric strain; this may disrupt the contact between A2 and A3 (important in continued association of A2 with the rest of the FVIIIa molecule following activation).
L412F (FVIII:C 7%, FVIII:Ag 6.4%; CRM−) is a mutation resulting in decreased expression or stability. The Leu residue is conserved in bovine and human FV, but is in fact replaced by the variant residue Phe in rat and human Cp. It is predicted to be partly buried with the sidechain oriented towards the core. When mutated to Phe in FVIII, a steric clash occurs with the S409 sidechain (the corresponding residue at this position in hCp is alanine). Expression or stability of the variant molecule is clearly affected by this change as the FVIII:Ag level is grossly reduced; however, functional activity is concordant with antigen level indicating that procoagulant function is unaffected in this variant.
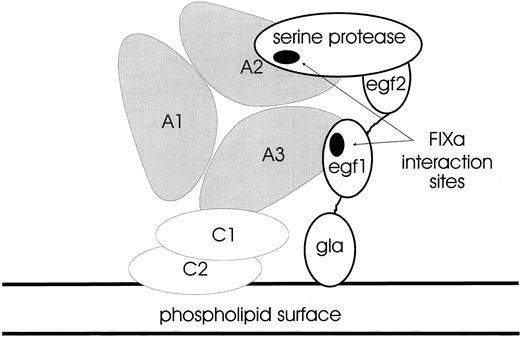
Hypothetical configuration of the FX-activating complex.It is plausible to orientate the triplicated A domains of FVIII/FVIIIa with a phospholipid surface via a specific interaction involving the C2 domain. Thus, the molecule might be held with the A3 domain closest to the surface: factors IXa and X also bind to such a surface via their N-terminal γ-carboxyglutamic acid-rich (Gla) domains. The EGF1 domain of FIXa has been shown to contact the A3 domain binding-site (E1811-K1818) whereas its protease domain contacts the A2 domain binding-site within S558-Q565; thus, the FIXa molecule may be aligned with the FVIII structure as in Fig 8. Since binding-site(s) for FX have not yet been identified, one can only speculate how the complex might interact with its substrate; however, there are potential surfaces for accommodation of substrate on the A1 domain and parts of A2. Alternatively, the cofactor function of activated FVIII may be explicable in terms of the orientation and allosteric activation of FIXa alone.
Hypothetical configuration of the FX-activating complex. A cartoon showing how the triangular array of the FVIII A1, A2, and A3 domains may be anchored to a phospholipid surface via the C1 and C2 domains (not to scale). The FIXa serine protease domain interacts with the loop in A2 (S558-Q565), whereas the FIXa EGF2 domain binds to the loop in A3 (E1811-K1818). The FIXa Gla domain is also anchored to the phospholipid surface via a calcium-dependent mechanism. This complex between FVIII and FIXa activates zymogen FX as a key step in the amplification of the initial trigger in coagulation.
Hypothetical configuration of the FX-activating complex. A cartoon showing how the triangular array of the FVIII A1, A2, and A3 domains may be anchored to a phospholipid surface via the C1 and C2 domains (not to scale). The FIXa serine protease domain interacts with the loop in A2 (S558-Q565), whereas the FIXa EGF2 domain binds to the loop in A3 (E1811-K1818). The FIXa Gla domain is also anchored to the phospholipid surface via a calcium-dependent mechanism. This complex between FVIII and FIXa activates zymogen FX as a key step in the amplification of the initial trigger in coagulation.
CONCLUSION
From the point of view of functional analysis of FVIII, it is unfortunate that circulating FVIII:Ag levels are only reported for a minority of missense mutation cases in the hemophilia A database. Only when these levels are reported is it possible to differentiate between the two likely causes of clinical disease resulting from missense mutations; either (1) reduced secretion/stability (low or absent FVIII:Ag, CRM−) or (2) dysfunction of a normally-secreted molecule (normal FVIII:Ag with reduced FVIII:C, CRM+). In the latter CRM+ class, hypotheses based on our model may be tested by performing site-directed mutagenesis studies to investigate, for example, FIXa interaction, thrombin-activation rate or stability of FVIIIa in recombinant variants.
Molecular models are not a substitute for high-resolution structures obtained by X-ray diffraction or NMR; however, in the absence of such structures for any part of FVIII or FVIIIa, we intend this model to point the way to a better understanding of the molecular pathology of hemophilia A, by facilitating the rational selection of experimental approaches including in vitro mutagenesis.
ACKNOWLEDGMENT
The authors thank A.I. Wacey (Thrombosis Research Institute, London, UK) for valuable initial assistance with construction of the model. The coordinates of the model in Protein Data Bank format, together with VRML (Virtual Reality Markup Language) representations, will be available following publication at the World Wide Web site of the Hemophilia A Database. URL: http://europium. mrc.rpms.ac.uk
Supported by the Medical Research Council and the Council for the Central Laboratories of the Research Councils.
Address reprint requests to S. Pemberton, PhD, Haemostasis Research Group, Medical Research Council Clinical Sciences Centre, Royal Postgraduate Medical School, Du Cane Rd, London W12 0NN UK.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal