Abstract
The heterodimeric complex glycoprotein (GP)IIb-IIIa, the fibrinogen receptor of platelets, carries numerous alloantigen systems. These polymorphisms are responsible for the immune response after transfusion or during pregnancy. In the latter case, the mother develops an antibody against an epitope present on fetal platelets, and this results in platelet destruction in the fetus. In this report, we describe the molecular characterization of a new alloantigen (Laa) on GPIIIa responsible for neonatal alloimmune thrombocytopenia (NAIT). Using polymerase chain reaction (PCR)–singlestrand conformation polymorphism (SSCP) and DNA sequencing, we found a point mutation (G to A) in a heterozygous state on the GPIIIa gene leading to amino acid substitution Arg to Gln at position 62 of the mature protein. Transient expression of GPIIb-IIIa complexes in Cos-7 cells using wild-type or mutated GPIIIa cDNA allowed us to demonstrate that this mutation was responsible for expression of the Laa epitope.
PLATELET ALLOIMMUNIZATION results from exposure to incompatible allotypic determinants. Platelet alloantigen systems are involved in the development of two well-described syndromes: fetal/neonatal alloimmune thrombocytopenia (NAIT) and posttransfusion purpura. NAIT was first described in the early 1950s,1 and is characterized by the destruction of fetal platelets by a maternal alloantibody elicited by fetomaternal incompatibility.2 NAIT is a transient passive disease in an otherwise healthy newborn, but there is a risk of intracerebral hemorrhage and therefore of neurologic impairment or death during the thrombocytopenic period. The incidence of NAIT has been recently estimated at one per 1,000.3 Currently, nine platelet alloantigen systems (human platelet antigen [HPA]) are established. Among the HPA systems giving rise to NAIT, the most frequently encountered in whites are HPA-1 and HPA-5, carried by the fibrinogen receptor (glycoprotein [GP]IIb-IIIa complexes) and the collagen receptor (GPIa-IIa complexes), respectively.
The GPIIb-IIIa receptor belongs to the integrin family and is responsible for platelet aggregation.5 This function is mediated through the binding of ligands containing the Arg-Gly-Asp (RGD) peptidic sequence6 such as von Willebrand factor, fibronectin, or vitronectin, whereas the binding of fibrinogen, the principal cofactor of aggregation, is mediated by the γ-chain dodecapeptide.7 Knowledge of the genes coding for GPIIb and GPIIIa8,9 has permitted definition of the HPA systems carried on these glycoproteins at the genetic level. It has been shown that GPIIb-IIIa is highly polymorphic and that different epitopes are due to single amino acid substitutions induced in each case by a point mutation on one or another of the genes. The genetic determinants of HPA-1, -4, -6, -7, and -8 are carried on GPIIIa.10-14 In contrast, HPA-315 and the recently described antigen Maxa16 are carried on GPIIb. The genetic determinant of Vaa is unknown, although the epitope is located on GPIIb-IIIa,17 as is the private system, Gro.18
We report here the genetic determination of a novel platelet alloantigen located on GPIIIa and responsible for a case of NAIT in a white family. The mutation was identified using a nonradioactive single-strand conformation polymorphism (SSCP) procedure and direct DNA sequencing. A single base pair substitution, G to A, at nucleotide 281 on GPIIIa cDNA, according to the published sequence,19 gave rise to an Arg to Glu amino acid substitution present in the heterozygous state at position 62 of mature GPIIIa. We then determined the alloantibody reactivity with recombinant antigens obtained through heterologous expression of wild-type and mutated GPIIb-IIIa in Cos-7 cells. This approach allowed us to determine that the mutation was responsible for expression of the epitope involved in the immune response and consequently in NAIT.
MATERIALS AND METHODS
Case report.We report a 29-year-old white woman (C.M.) who gave birth to her first child (a girl); petechiae were observed 1 hour after birth, and after 4 hours the infant was admitted to the intensive care unit. Otherwise, the initial physical examination was normal. The platelet count showed a severe thrombocytopenia with 19 × 109 platelets/L, and this was decreased to 4 × 109/L 10 hours later. The infant was transfused with washed maternal platelets obtained by apheresis and treated with intravenous IgG 1 g/kg/d for 2 days. The outcome was favorable; the platelet count had increased to 454 × 109/L on day 6, when the child was discharged from the hospital.
Blood samples.Blood samples from all available family members, including the father (L.G.), and healthy adult human volunteers were collected by venipuncture using 10 mmol/L EDTA as an anticoagulant (9 vol blood:1 vol anticoagulant). The child was not made available for study.
Platelet serology.Platelet phenotyping and characterization of maternal serum alloantibodies were performed with the monoclonal antibody (MoAb)-specific immobilization assay (MAIPA) as previously described.20 The following MoAbs were used: GR-P against GPIX (from Dr Garrido, Granada, Spain); Gi9 against GPIa (from Dr Santoso, Giessen, Germany); P91 against β2-microglobulin (Fourth International Workshop on Leukocyte Typing, 1988); and P11-64 against GPIIb and P12-46 against GPIIb-IIIa (from the Institut National de la Transfusion Sanguine [INTS], Paris, France). Platelet phenotyping was performed using the following alloantisera: anti–HPA-1a (IgG), anti–HPA-1b (IgG), anti–HPA-3a (IgG), anti–HPA-4a (IgG), anti–HPA-4b (IgG), anti–HPA-5a (IgG), anti–HPA-5b (IgG), anti–HPA-6b (anti-Tua; typing performed by Dr Kekomaki, Helsinki, Finland), anti–HPA-7b (anti-Moa; from Dr von dem Borne, Amsterdam, The Netherlands), anti–HPA-8b (anti-Sra; from Dr Kiefel, Giessen, Germany), anti-Vaa (IgG; typing performed by Dr Kekomaki), and anti-Groa (IgG; from Dr von dem Borne).
Enzymatic treatment of platelets and localization of the epitope.Paternal platelets were treated with α-chymotrypsin for 30 minutes at 37°C as already described,21 and the reaction was stopped by addition of phenylmethylsulfonyl fluoride (PMSF). After centrifugation, the supernatant was treated with 2% (wt/vol) sodium dodecyl sulfate (SDS) and boiled for 10 minutes. The platelet pellet was resuspended in 10 mmol/L Tris hydrochloride, pH 7.4, 150 mmol/L NaCl, 3 mmol/L EDTA, 2% (wt/vol) SDS, 2.5 mmol/L PMSF, and 30 mmol/L N-ethylmaleimide and boiled for 10 minutes. Western blot experiments were performed after SDS–polyacrylamide gel electrophoresis (SDS-PAGE) under nonreduced and reduced conditions as previously described.22
Isolation of genomic DNA.Genomic DNA from available family members and unrelated donors was isolated from peripheral blood according to published procedures.23
Polymerase chain reaction–SSCP analysis, DNA sequencing, and allele-specific restriction analysis (ASRA).To determine which exon carried the mutation responsible for the new epitope, we performed SSCP analysis24 on the exons of GPIIIa coding for the chymotrypsin-insensitive part of GPIIIa that was responsible for the binding of antibodies in the serum of the mother. For DNA amplification, we used different pairs of oligonucleotides and reaction conditions that have been previously described.25 Each amplification product contained one exon, an exception being exon 10, which was amplified with two different pairs of oligonucleotides to produce appropriate-sized DNA fragments, 10(1) and 10(2), for SSCP analysis. Nonradioactive SSCP analysis was performed using the PhastSystem minigel electrophoresis system (Pharmacia-Biotech, Orsay, France).26 Sequencing of polymerase chain reaction (PCR) products was performed using the fmol DNA Sequencing System (Promega, Lyon, France) and specific primers radiolabeled with 33P (Isotopchim; Ganagobie-Peyruis, France) according to the conditions recommended by the manufacturers. As the described mutation abolished a recognition site for the restriction enzyme Ava I, this enzyme was used to assess the presence of the mutation in DNA from family members and unrelated individuals according to the conditions recommended by the manufacturer (New England Biolabs, Beverly, MA).
Site-directed mutagenesis and expression in Cos-7 cells.The transient eucaryotic expression vector pSM was constructed from pSG5 (Stratagene, La Jolla, CA) with the insertion between the unique EcoRI and BglII restriction sites of a synthetic double-strand DNA fragment bearing a new cloning multisite (5′EcoRI-Pst IHindIII-BamHI-BglII3′). The full-length cDNA encoding wild-type GPIIb was excised from pIPNDIIb (a generous gift from Prof Paul Bray, Johns Hopkins University, Baltimore, MD) by total digestion with both EcoRI and HindIII restriction enzymes, and subcloned into EcoRI/HindIII-digested pSM. The full-length cDNA encoding wild-type GPIIIa was excised from pIBIIIa (from Prof Bray) by partial digestion with EcoRI and total digestion with Hind III restriction enzymes, and subcloned into pAlter (Promega). The internal EcoRI restriction site of GPIIIa cDNA was subjected to site-directed mutagenesis without changing coding properties using the Altered Sites In Vitro Mutagenesis System (Promega). The mutation that we found was introduced into wild-type GPIIIa cDNA and oligonucleotide 5′GAGGCCCAAGTACTAGAGG3′, where the underlined A replaced a G in the wild-type sequence.19 As this mutation abolished the restriction sequence recognized by Ava I restriction enzyme, we checked for the mutant using this enzyme. The entire cDNA of the mutated clones was sequenced to ensure that no other mutation was introduced during the site-directed mutagenesis experiments. Mutated cDNA was then subcloned into pSM as described earlier.
Cos-7 cells (a generous gift from Dr A. Sarasin, IRSC, Villejuif, France) were cultivated in MEM medium (GIBCO-BRL, Life Technologies, Cergy-Pontoise, France) supplemented with 7% fetal calf serum (D. Dutscher, Brumath, France) in a 5% CO2 saturating-humidity atmosphere. Forty-eight hours before transfection, 2 × 106 Cos-7 cells were seeded on 10-cm petri dishes. For transfection, 6 μg of each plasmid was mixed with Transfectam reagent (Promega) and placed in contact with the cells according to the conditions given by the manufacturer. After 4 hours at 37°C, 15 mL complete medium was added to each petri dish and the cells were incubated for 48 to 76 hours before analysis.
Analysis for the Presence of the Epitope on Recombinant GPIIb-IIIa Molecules
Transfected cells were harvested as previously described,27 counted, and divided into two parts before being analyzed for GPIIb-IIIa expression.
Flow cytometry.Cells (400,000 for each analysis) were resuspended in 0.5 mL phosphate-buffered saline containing 0.1% (wt/vol) bovine serum albumin (PBS-BSA) and 3 μg/mL EDU3, a murine MoAb specific for the GPIIb-IIIa complex (a generous gift from Dr R. Vilella, Barcelona, Spain), and incubated at room temperature for 30 minutes. After sedimentation at 300 g for 10 minutes and washing with PBS-BSA, cells were incubated for 30 minutes at room temperature in 0.5 mL PBS-BSA containing FITC-labeled F(ab)2 antibody to mouse IgG (Silenus Laboratories, Hawthorn, Australia). After another round of centrifugation and washing, cells were analyzed using a Becton Dickinson FACScan (Becton Dickinson, Le Pont de Claix, France).
Detection of recombinant antigens on transfected Cos-7 cells.Expression of the recombinant GPIIIa epitope was analyzed by a method derived from the MAIPA.20 Collected transfected cells were resuspended in 160 μL PBS-BSA supplemented with 40 μL nonimmune serum, serum from patient E.V., a Glanzmann's thrombasthenia patient who possesses a well-characterized antibody to GPIIIa,28 or serum from patient C.M. After 14 hours at 4°C, cells were sedimented as described earlier and resuspended in 200 μL PBS-BSA containing 10 μg IgG from clone Y2/51, an MoAb to GPIIIa (Dakopatts, Glostrup, Denmark). After 1 hour at 37°C, cells were centrifuged and resuspended in 50 μL H2O and lysed following addition of 50 μL 20-mmol/L Tris hydrochloride, pH 7.5, 10 mmol/L EDTA, 2% (wt/vol) Nonidet P-40, and 2 mmol/L PMSF. After 30 minutes at 4°C, the lysate was centrifuged for 30 minutes at 12,000g and the supernatant was harvested. ELISA plates were treated under the conditions described for the MAIPA test.20 For each analysis, 100 μL cell lysate was loaded in triplicate, and plates were incubated for 90 minutes at 4°C. After six washes with PBS containing 0.05% (vol/vol) Tween 20 (Sigma) and 2% (wt/vol) nonfat dry milk powder (washing buffer), bound human antibody was detected following addition of 100 μL per well of goat anti–human IgG and IgM conjugated to alkaline phosphatase enzyme (Jackson ImmunoResearch, West Grove, PA) diluted 1:1,000. After incubation for 1 hour at room temperature, wells were washed six times with washing buffer, and 200 μL 2-mg/mL p-nitrophenyl phosphate (Sigma) in 1 mol/L diethanolamine and 0.5 mmol/L MgCl2 , pH 9.8, was added. Absorbances were read at 405 nm after 16 hours at 20°C in the dark.
RESULTS
Serologic studies.We began by phenotyping platelets from the mother (C.M.), the father, and the child's paternal grandparents for each of the known alloantigen systems for which the appropriate antiserum was available. The results showed no potential incompatibility within these systems. We next determined the glycoprotein against which the maternal antibody was directed by performing a series of MAIPA using MoAbs specific for each of the major membrane glycoproteins of platelets. The maternal serum gave a selective and strong positive reaction on the father's and paternal grandmother's platelets with Pl1-64 and Pl2-46 (MoAbs that recognize different determinants on the intact GPIIb-IIIa complex on platelets). Results were negative with the other MoAbs tested (MoAbs to GPIb-IX, Ia-IIa complex, and HLA class I antigens; data not shown). Results were negative for all MoAbs with platelets from the mother or the paternal grandfather. Overall, these results suggested that the antibody present in the mother's serum was directed against an undescribed epitope, which we have called Laa, carried on the GPIIb-IIIa complex. Furthermore, the maternal serum did not react with platelets from a panel of 60 typed and unrelated donors. We therefore proceeded to locate this new alloepitope and to characterize its genetic determinant.
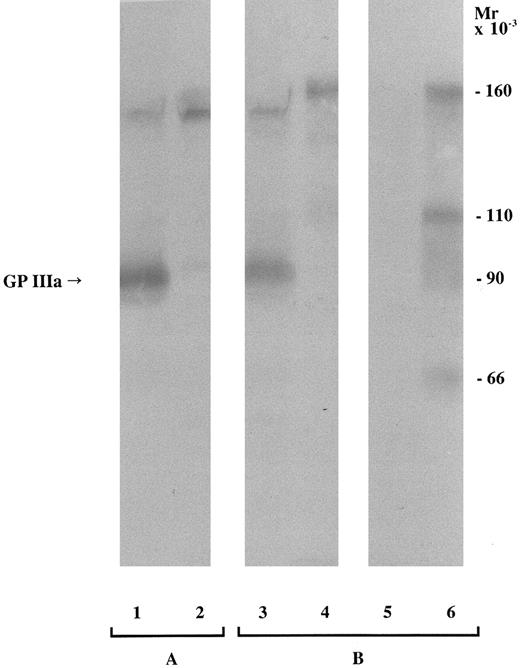
Immunochemical characterization.Since the alloantibody appeared to be directed against an epitope on GPIIb-IIIa, we performed Western blotting experiments to identify the subunit that carried the epitope. Maternal serum contained an antibody that bound specifically to a protein migrating under nonreduced conditions in the position of GPIIIa of the father's platelets (Fig 1, lane 3). After reduction, a band was again observed at a molecular mass corresponding to GPIIIa after disruption of disulfide bonds, although its intensity was now weaker (data not shown). It has been demonstrated that treatment of platelets with α-chymotrypsin leads to digestion of GPIIIa giving rise to three polypeptides that represent amino acids 1 to 100, 101 to 321, and 322 to 762, respectively.29,30 Polypeptides constituted by amino acids 1 to 100 and 322 to 762 are disulfide-linked and anchored in the platelet membrane.29 Thus, after centrifugation of α-chymotrypsin–treated platelets, the polypeptide corresponding to amino acids 101 to 321 remains in the supernatant while the other polypeptides are retained in the platelet membrane and consequently are present in the platelet pellet. When platelets from the father were treated with α-chymotrypsin, the epitope carried on GPIIIa was present only in the pellet (Fig 1, lane 6). A band observed at 110 kD corresponds to a polypeptide incompletely digested by α-chymotrypsin, and a band at 66 kD to the disulfide-linked polypeptides retained in the membrane.28-30 It was therefore concluded that the epitope recognized by the mother's antibody was located on the extracellular domain of GPIIIa but outside amino acids 101 to 321.
Immunoblot of SDS-soluble platelet proteins subjected to SDS-PAGE (6.5% polyacrylamide gel) under nonreduced conditions and transferred to nitrocellulose membrane. For each sample, 150 μg total platelet protein was loaded. An anti–HPA-1a serum was used as a positive control (A), and the mother's serum was tested (B). Lanes: (1) HPA-1–positive platelets, (2) HPA-1–negative platelets, (3) father's platelets, (4) mother's platelets, (5) supernatant recovered after α-chymotrypsin treatment and sedimentation of the father's platelets, and (6) pellet after α-chymotrypsin treatment and centrifugation of the father's platelets. The band at ≈160 kD represents platelet-associated IgG.
Immunoblot of SDS-soluble platelet proteins subjected to SDS-PAGE (6.5% polyacrylamide gel) under nonreduced conditions and transferred to nitrocellulose membrane. For each sample, 150 μg total platelet protein was loaded. An anti–HPA-1a serum was used as a positive control (A), and the mother's serum was tested (B). Lanes: (1) HPA-1–positive platelets, (2) HPA-1–negative platelets, (3) father's platelets, (4) mother's platelets, (5) supernatant recovered after α-chymotrypsin treatment and sedimentation of the father's platelets, and (6) pellet after α-chymotrypsin treatment and centrifugation of the father's platelets. The band at ≈160 kD represents platelet-associated IgG.
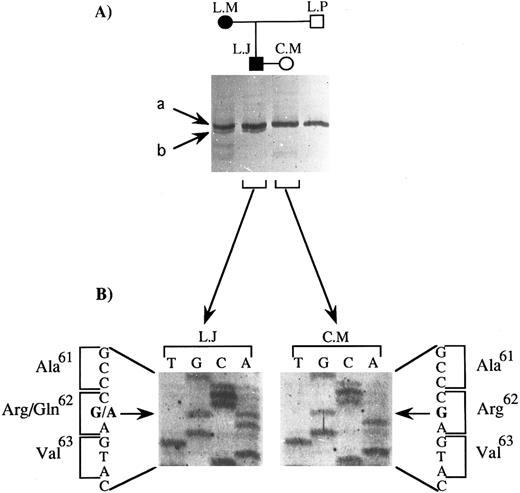
Determination of the mutation.We analyzed the portion of the GPIIIa gene coding for the extracellular domain of the fragments of GPIIIa that remained associated with platelets after α-chymotrypsin treatment. According to Zimrin et al19 and Villa-Garcia et al,31 amino acids 1 to 100 and 322 to 692 (amino acids 693 to 762 not being in the extracellular domain of GPIIIa) are coded for by exons 2 to 4 and 8 to 14, respectively. We performed a nonradioactive SSCP analysis of the amplification products from DNA obtained from the two parents and the two paternal grandparents. We recall that the epitope was present on both the father's and the paternal grandmother's GPIIIa. DNA from the affected child was not available for study. Only the migration profile of PCR products containing exon 3 showed differences between family members. This is illustrated in Fig 2A. Two bands, a and b, were observed for the father and paternal grandmother (L.J. and L.M., respectively), while only band a was seen for the mother and paternal grandfather (C.M. and L.P., respectively). This difference could not be explained by differences in the HPA-1 system, for which the genetic determinant is carried by the same exon, serologic analysis having shown that all four family members were homozygous for HPA-1a. Moreover, the SSCP pattern that we observed for C.M. and L.P. was strictly identical to that previously described by our group for this exon for individuals homozygous for HPA-1a, whereas the SSCP pattern for L.J. and L.M. for this exon did not correspond to that characteristic for HPA-1a/1b or HPA-1b/1b.26 Sequencing of the PCR product containing exon 3 from the two parents (Fig 2B) revealed a single base substitution present in a heterozygous state in DNA from the father (G to A at nucleotide 281), whereas the DNA sequence of exon 3 of the mother was exactly as published.19 This mutation leads to an amino acid substitution (Arg to Gln) at position 62 on mature GPIIIa.
SSCP analysis and sequencing of exon 3 of the GPIIIa gene: identification of the mutation carried by the father (L.J.). (A) SSCP analysis of exon 3 of 4 family members. Two bands, a and b, were seen for L.J. and L.M., for whom C.M.'s serum was reactive against their platelets, while only band a was observed for C.M. and L.P., for whom C.M.'s serum was unreactive against their platelets. (B) Direct DNA sequencing of the corresponding amplification product for L.J. and C.M. The position of the mutation is indicated with a horizontal arrow, and the substituted base with a bold letter; the amino acid substitution is indicated together with its position in the GPIIIa protein.
SSCP analysis and sequencing of exon 3 of the GPIIIa gene: identification of the mutation carried by the father (L.J.). (A) SSCP analysis of exon 3 of 4 family members. Two bands, a and b, were seen for L.J. and L.M., for whom C.M.'s serum was reactive against their platelets, while only band a was observed for C.M. and L.P., for whom C.M.'s serum was unreactive against their platelets. (B) Direct DNA sequencing of the corresponding amplification product for L.J. and C.M. The position of the mutation is indicated with a horizontal arrow, and the substituted base with a bold letter; the amino acid substitution is indicated together with its position in the GPIIIa protein.
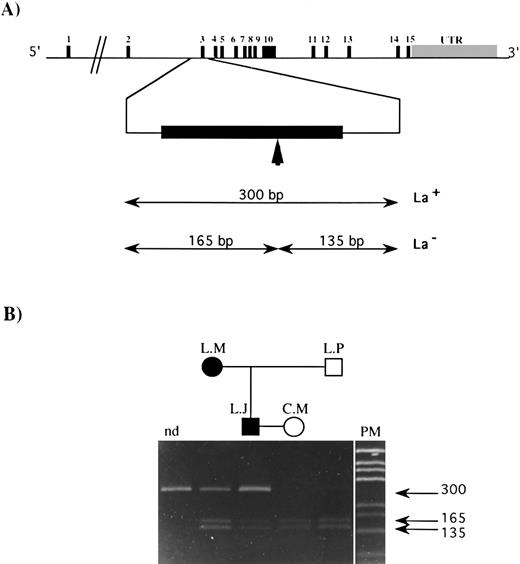
The observed mutation abolished the unique sequence for the restriction enzyme AvaI on this part of the DNA. The GPIIIa gene structure and the size of the amplification product containing exon 3 and that of the DNA fragments obtained after digestion with AvaI, both when the mutation was present (mutated type) or absent (wild type), are indicated in Fig 3A. The result of the allele-specific restriction analysis (ASRA) on this PCR product from the family members (Fig 3B) demonstrates clearly that L.M. and L.J. are heterozygous for this mutation, while C.M. and L.P. are homozygous for the wild-type sequence. This mutation has never been reported in the literature and has not been found by our group in DNA from a series of 60 independent donors using SSCP or ASRA.
ASRA on exon 3 of the GPIIIa gene from family members. (A) Genomic organization of the GPIIIa gene. The 15 exons are symbolized by vertical black bars, and the exon number is indicated; UTR refers to the untranslated region. The vertical arrow indicates the target sequence recognized by the Ava I restriction enzyme in the published sequence.19 The size of restriction fragments resulting from Ava I digestion of exon 3 for the La genotype is indicated (La+, mutated type; La−, wild-type). (B) A 10% polyacrylamide gel on which was loaded amplification products containing exon 3 of the gene for GPIIIa: nd, undigested amplification product from a normal donor, amplification products from L.M., L.J., C.M., and L.P. digested with the Ava I restriction enzyme. PM, pGEM DNA Markers (Promega). The size of each DNA fragment, in base pairs, is shown by horizontal arrows.
ASRA on exon 3 of the GPIIIa gene from family members. (A) Genomic organization of the GPIIIa gene. The 15 exons are symbolized by vertical black bars, and the exon number is indicated; UTR refers to the untranslated region. The vertical arrow indicates the target sequence recognized by the Ava I restriction enzyme in the published sequence.19 The size of restriction fragments resulting from Ava I digestion of exon 3 for the La genotype is indicated (La+, mutated type; La−, wild-type). (B) A 10% polyacrylamide gel on which was loaded amplification products containing exon 3 of the gene for GPIIIa: nd, undigested amplification product from a normal donor, amplification products from L.M., L.J., C.M., and L.P. digested with the Ava I restriction enzyme. PM, pGEM DNA Markers (Promega). The size of each DNA fragment, in base pairs, is shown by horizontal arrows.
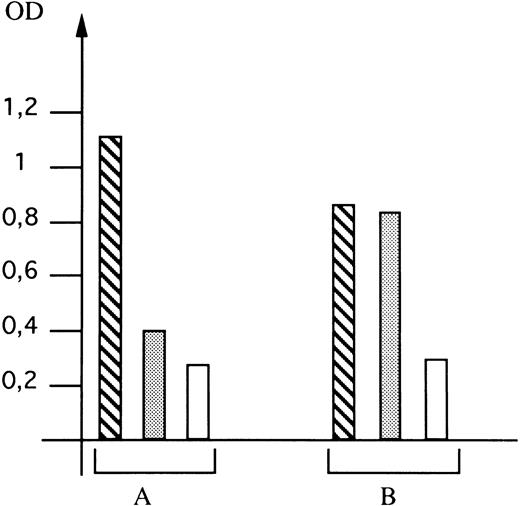
Expression of the mutated GPIIIa in Cos-7 cells.The direct involvement of the G to A mutation at nucleotide 281 of GPIIIa cDNA in the formation of the epitope recognized by the mother's serum was investigated by cotransfecting Cos-7 cells with eucaryotic expression vectors carrying wild-type cDNA for GPIIb and wild-type or mutated cDNA for GPIIIa. Transfection experiments were performed under transient expression conditions, the percentage of cells with GPIIb-IIIa complexes at the surface being determined by flow cytometry performed using the MoAb EDU3, which recognizes a complex-dependent determinant on GPIIb-IIIa. Data from three independent experiments showed that a mean of 9.8% and 10.1% of cotransfected cells expressed the complex when cotransfection was performed with wild-type cDNA for GPIIb and wild-type or mutated cDNA for GPIIIa, respectively (data not shown). Since this mutation did not alter the cell membrane expression of GPIIb-IIIa, we used different sera to determine if its presence led to the formation of the epitope recognized by the antibody present in C.M.'s serum. For this, we developed an assay that permits detection of the presence of a particular epitope on recombinant GPIIb-IIIa without using radioactivity and when the epitope is not detectable in Western blotting experiments (as is the case for complex-dependent determinants). The results are shown in Fig 4. When a nonimmune serum was used, only background binding was observed, whatever GPIIIa cDNA had been used. As positive control, we used serum from patient E.V., who had developed an isoantibody to GPIIIa subsequent to blood transfusion.28 Binding of this antibody to the GPIIb-IIIa complex was observed using cells transfected with both wild-type and mutated GPIIIa. In contrast, when serum from C.M. was used, significant binding was only observed when mutated GPIIIa was present. This result clearly demonstrated that the mutation was directly involved in the expression of the Laa epitope on GPIIIa.
Detection of expression of the Laa epitope on recombinant GPIIb-IIIa complexes. Expression on Cos-7 cells of GPIIb-IIIa complexes containing wild-type (A) or mutated (B) GPIIIa was determined using E.V.'s serum as a positive control (▧), nonimmune serum as a negative control (□), and C.M.'s serum (▧). The optical density (OD) was read 16 hours after addition of the chromogenic substrate. Values are the mean of 6 points obtained in two different experiments. The same number of cells were analyzed for each serum tested.
Detection of expression of the Laa epitope on recombinant GPIIb-IIIa complexes. Expression on Cos-7 cells of GPIIb-IIIa complexes containing wild-type (A) or mutated (B) GPIIIa was determined using E.V.'s serum as a positive control (▧), nonimmune serum as a negative control (□), and C.M.'s serum (▧). The optical density (OD) was read 16 hours after addition of the chromogenic substrate. Values are the mean of 6 points obtained in two different experiments. The same number of cells were analyzed for each serum tested.
DISCUSSION
We report here the identification and characterization of a new platelet alloantigen, Laa, involved in a case of NAIT. The question of whether it is of low frequency or a private alloantigen confined to the father's family remains to be answered. In our studies, the Laa determinant was subsequently not detected in a panel of 60 individuals. Cleavage of GPIIIa by α-chymotrypsin revealed that the determinant was located outside the amino acid sequence 101 to 321 of GPIIIa. PCR-SSCP analysis of exons of the GPIIIa gene coding for the sequences outside amino acids 101 to 321 yet extracellular in position revealed migration changes in exon 3. After sequencing the amplified PCR product of exon 3, we found a single point mutation (G to A at nucleotide 281) present in a heterozygous state in the father's genome, and ASRA additionally showed that it was present in the paternal grandmother's GPIIIa gene. This mutation leads to amino acid substitution Arg to Gln at position 62 on mature GPIIIa. Finally, expression of recombinant mutated GPIIIa in Cos-7 cells led to the confirmation that this single amino acid substitution was sufficient to induce formation of the epitope recognized by the antibody present in the mother's serum. Newman32 has recently proposed an alternative nomenclature for human platelet alloantigens based on the fact that most polymorphisms arise through mutations of the same wild-type allele. It is important to emphasize that in our family, the Laa epitope is given by the Gln62 polymorphism, while the alternative allele corresponds to that giving rise to the GPIIIa sequence reported by Zimrin et al19 and has a gene frequency of 0.85.32
Western blotting established that binding of maternal antibody to the Laa determinant was lower after reduction of disulfide bonds, but not abolished. In this way, it differs from HPA-1a antibodies, which do not recognize disulfide-reduced GPIIIa. This difference is interesting in view of the closeness of the Laa epitope to the HPA-1 system, which is located on amino acid 33 of GPIIIa.8 Two categories of anti–HPA-1a antibodies have been defined: (1) those for which only an intact amino terminus is required for binding, and (2) those for which binding is dependent on other structural requirements within the entire glycoprotein.33 Nevertheless, the disruption of the Cys26-Cys38 loop of GPIIIa abolished the binding of all anti–HPA-1a alloantibodies tested in previous studies.33,34 Although we cannot totally exclude that some GPIIIa molecules in our study were incompletely reduced, in particular for the Cys26-Cys38 loop, this is unlikely, since we observed a single band in the position of fully reduced GPIIIa (data not shown). Thus, another explanation is that the binding of the mother's antibody is not completely under the dependence of this loop, but occurs to reduced GPIIIa with a lower affinity. Binding experiments using recombinant GPIIIa molecules with a disrupted Cys26-Cys38 loop will be of interest to clarify this point. Furthermore, site-directed mutagenesis involving the Laa alloantigen and the HPA epitopes could be an interesting way to explore the structural requirement of the NH2-terminus of GPIIIa in the humoral response. Interestingly in this regard, a nonalloantigenic polymorphism at residue 40 has also been described.35
Recently, a mutation in the codon for Arg62 of GPIIIa leading to type I Glanzmann's thrombasthenia has been reported.36 In this case, the mutation C to T (or G to A on the other strand) is located in a CpG dinucleotide and leads to a stop codon inducing a truncated protein unable to form a functional heterodimer. In this context, our result clearly demonstrates that the presence of Arg62 is not obligatory for the expression of the GPIIb-IIIa complex at the cell surface, since an Arg62 to Gln substitution has no effect on this expression. The codon for Arg62 thus seems particularly prone to mutagenesis. This can be correlated with the presence of a CpG at this site, this dinucleotide often being associated with point mutations.37 Interestingly, the HPA-6 system12 located on GPIIIa is encoded by the same genetic determinant: codon CGG (Arg489 ) for HPA-6a to codon CAG (Gln489 ) for HPA-6b where the mutation G to A is in a CpG dinucleotide. The mutation and amino acid substitution are the same in the two cases, and both give rise to NAIT. It is of note that the lateral chain of Arg is extremely basic and that of Gln is neutral; this point could be of importance in epitope formation.
The reason that the NH2 extremity of GPIIIa is highly polymorphic could be linked to the fact that substitutions in this region do not appear to lead to a loss of function in the receptor, at least in terms of platelet function testing as it is currently performed. In contrast, the RGD binding site6 and the sequence between amino acid 210 to 22138 are domains of GPIIIa recognized to be involved in fibrinogen binding to the GPIIb-IIIa complex. The key role of these regions has been reinforced by the description of amino acid substitutions found in patients with variant forms of Glanzmann's thrombasthenia and that lead to a loss in fibrinogen binding.39 Looking at the distribution of HPA systems carried by the GPIIb-IIIa complex, it is interesting that seven of them are located on GPIIIa, spanning the extracellular domain of the protein, while only two are located on GPIIb and more precisely close to the C terminus of the heavy chain. Taking into account this point and the fact that only the HPA-4 system coincides with a known functional domain,11 it is tempting to speculate that a requirement for the survival of alloantigen systems is the fact that the polymorphism allows at least a partial functioning of the receptor.
ACKNOWLEDGMENT
We thank C. Dabanian for technical assistance. We also thank Prof Paul Bray for help in initiating the PCR-SSCP procedure in Bordeaux.
Supported by the CNRS, Université de Bordeaux II, the Conseil Régional d'Aquitaine, the Fondation pour la Recherche Médicale, and the Ministère de l'Enseignement Supérieur et de la Recherche (Action Concertéc Coordonńea-Sciences de la Vie N. 9). O.P. is a recipient of a postdoctoral fellowship from the Société Française d'Hématologie.
Address reprint requests to F. Bourre, PhD, UMR CNRS 5533, Hôpital Cardiologique, Avenue de Magellan, 33604 Pessac, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal