Abstract
L-selectin is a leukocyte cell-surface glycoprotein that mediates adhesive interactions between circulating cells and vascular endothelium. All endothelial ligands of L-selectin characterized to date are glycoproteins that require sulfation for activity and share reactivity with MECA 79, a monoclonal antibody that recognizes a sulfate-dependent epitope involved in L-selectin attachment. We have recently identified by functional assay a glycoprotein L-selectin ligand expressed on the human hematopoietic cell line KG1a. We report here that this ligand is not recognized by MECA 79 and that it retains binding activity after metabolic inhibition of sulfation by chlorate. A native membrane L-selectin ligand exhibiting sulfate-independent function has not been described previously. Identification of this novel ligand on a nonendothelial cell type suggests that structural determinants conferring L-selectin binding may vary in a cell- and tissue-specific manner.
L-SELECTIN (CD62L) IS A leukocyte membrane glycoprotein that functions as a Ca2+-dependent lectin in adhesive interactions with endothelial membrane glycoprotein ligands. These interactions result in initial tethering and rolling of leukocytes on endothelium, critical steps in the directed migration of leukocytes to sites of inflammation and the physiologic trafficking of lymphocytes into peripheral lymph nodes. L-selectin was first identified operationally through observations of the binding of lymphocytes of rodents and humans to lymph node high endothelial venules (HEV).1,2 These investigations used an in vitro binding assay (the Stamper-Woodruff lymphocyte-HEV adherence assay) in which lymphocytes specifically adhere under shear conditions to frozen sections of lymph node HEV. The lymphocyte membrane structure mediating binding was initially identified by monoclonal antibodies (MoAbs) against mouse3 and rat4 lymphocytes that blocked binding to HEV in the Stamper-Woodruff assay. These antibodies subsequently led to the characterization of L-selectin at a molecular level, and full-length cDNA clones have now been characterized and sequenced in a variety of mammals, including humans,5-8 mice,9,10 and rats.11,12 In addition, the adherence assay facilitated the identification of the HEV L-selectin ligands; a rat antimurine lymph node MoAb was raised that specifically stains nodal HEV and blocks L-selectin–dependent lymphocyte adherence to HEV in the assay, as well as in vivo.13 This antibody, designated MECA 79, immunoprecipitates HEV L-selectin ligands (known as lymph node addressins) from the lymph node of both mouse and human, attaching in each case to a common epitope involved in functional adherence to L-selectin.
Most of what is known about L-selectin ligands has been derived from studies of molecules recognized by MECA 79. In murine lymph nodes, MECA 79 immunoprecipitates two sulfated glycoproteins of molecular weight (MW) 50,000 and 90,000, initially designated Sgp50 and Sgp90, respectively.14 A chimeric murine L-selectin-human IgG molecule (designated LEC-IgG) has been constructed that also immunoprecipitates the Sgp50 and Sgp90 proteins from lymph node lysates,14,15 and recent data suggest that a third glycoprotein of MW 200,000 is also recognized by MECA 79 and the chimera.16 In addition, the MECA 79 determinant is found on a selectively modified subset of the HEV glycoprotein known as the mucosal vascular addressin (MAdCAM-1), which also supports L-selectin–mediated lymphocyte adhesion under shear.17 The Sgp50 protein, now known as GlyCAM-1, has been cloned and its cDNA encodes a secreted protein consisting of 132 amino acids with multiple potential O-linked glycosylation sites.15 The Sgp90 protein is an integral membrane protein that appears to be an endothelial glycoform of CD34.18
The HEV ligands recognized by MECA 79 are heavily glycosylated mucin-like proteins rich in O-linked glycosylations bearing sialic acid. Treatment of endothelial L-selectin ligands with neuraminidase16,19 and with O-sialoglycoprotease20 abrogates the binding of L-selectin. Metabolic studies using chlorate as an inhibitor of sulfation have shown that the molecular determinant recognized by both MECA 79 and L-selectin chimera is sulfation-dependent,16,21 indicating that sulfation is critical to the function of native endothelial L-selectin ligands. Furthermore, the importance of sulfation in ligand activity has been emphasized by the finding that naturally expressed unsulfated GlyCAM-1 is not capable of binding L-selectin.22
We recently reported23 that the primitive human hematopoietic cell line KG1a24 expresses a functional L-selectin ligand, the first description of a nonendothelial, native membrane L-selectin ligand. This ligand was identified using a modification of the original Stamper-Woodruff assay in which cyto-spin preparations of cells substituted for lymph node sections. In these experiments, rodent and human lymphocytes each bound to KG1a under identical shear assay conditions in which L-selectin mediates lymphocyte binding to HEV in lymph nodes. The adherence of lymphocytes to KG1a was L-selectin–specific; binding was calcium-dependent, was strictly inhibited by anti–L-selectin antibodies and by carbohydrates known to bind L-selectin (eg, fucoidin), and was completely abrogated after phorbol ester-induced shedding of L-selectin from the lymphocyte membrane.23 Although KG1a cells express CD34 (which on hematopoietic cells serves as a marker of early progenitor cells including the stem cell population25 ), a variety of independent investigative approaches showed that CD34 as expressed on KG1a was not the ligand. Moreover, CD34 expressed on other human hematopoietic cell lines (including RPMI 8402 and MO7e) did not exhibit L-selectin ligand activity,23 indicating that hematopoietic CD34 (in contrast to CD34 of HEV) is not an L-selectin ligand.
Flow cytometric analysis of MECA 79 antigen on KG1a cells. Representative flow cytometric profiles are shown of (A) rat IgM isotype control and (B, bold line) MECA 79 primary antibodies, followed by FITC-conjugated secondary antibody. Note that MECA 79 levels were indistinguishable from isotype control.
Flow cytometric analysis of MECA 79 antigen on KG1a cells. Representative flow cytometric profiles are shown of (A) rat IgM isotype control and (B, bold line) MECA 79 primary antibodies, followed by FITC-conjugated secondary antibody. Note that MECA 79 levels were indistinguishable from isotype control.
Enzymatic studies showed that the KG1a L-selectin ligand is a glycoprotein and that L-selectin binding activity is sialic acid-dependent (ie, eliminated by neuraminidase treatment), but, unlike endothelial L-selectin ligands, the KG1a ligand is resistant to digestion by O-sialoglycoprotease.23 This differential sensitivity of native membrane ligands from endothelium and from KG1a to O-sialoglycoprotease digestion raised the hypothesis that functional L-selectin binding determinants may differ among ligands in a cell-specific fashion. In the present study, we explored this hypothesis by directly investigating whether KG1a cells express MECA 79 antigen(s) and by examining the role of sulfation in KG1a L-selectin ligand activity. We report here that the KG1a L-selectin ligand does not contain MECA 79 antigenic epitopes and that sulfation is not critical to its function. These data provide first evidence of sulfate-independent recognition of L-selectin by a naturally expressed, mammalian L-selectin ligand and establish that L-selectin binding determinants are not structurally conserved among native membrane glycoprotein L-selectin ligands expressed on different cell types.
MATERIALS AND METHODS
Cells and antibodies.KG1a cells were cultured in RPMI 1640 (GIBCO-BRL, Gaithersburg, MD) containing 10% fetal bovine serum (FBS) in a humidified chamber at 37°C with 5% CO2. MoAb MECA 79 and rat IgM-isotype control MoAb were gifts of Dr P. Streeter (Searle Research Laboratories/Monsanto Co, St Louis, MO). Unconjugated anti-CD43 (leukosialin) MoAb L60 and isotype (murine IgG1 ) control were obtained from Becton Dickinson (San Jose, CA). Fluorescein isothiocyanate (FITC)-conjugated goat antirat IgM and phycoerythrin (PE)-conjugated goat antimouse IgG antibodies were obtained from Jackson ImmunoResearch Labs Inc (West Grove, PA).
Immunofluorescence studies.Indirect immunofluorescence staining for MECA 79 antigens was performed on acetone-fixed KG1a cytospins, on acetone-fixed murine peripheral lymph node sections (to assess HEV staining as controls), and on unfixed KG1a cell suspensions. KG1a cytospins, murine lymph node sections, and KG1a suspensions (1 × 106 KG1a in 100 μL phosphate-buffered saline [PBS] containing 2% FBS on ice) were in each case incubated with 1 μg MECA 79 or IgM control antibody in PBS/2% FBS for 30 minutes. After washing in PBS/2% FBS, the cells or sections were treated with 5 μg FITC-conjugated goat antimouse IgM secondary antibody, followed by additional washes in PBS/2% FBS. MECA 79 staining of KG1a cytospins and of lymph node HEV was assessed by fluorescence microscopy. Analysis of MECA 79 expression on KG1a cell suspensions was performed using flow cytometry. Flow cytometric analysis of CD43 on KG1a was performed using PE-conjugated secondary antibody after primary anti-CD43 antibody L60 or isotype control antibody incubation as per the manufacturer's recommendations. Flow cytometry was performed on a FACStarPLUS (Becton Dickinson) apparatus.
Metabolic radiolabeling and immunoprecipitation.KG1a were metabolically radiolabeled by incubating 108 cells (20 × 106 cells/mL) for 8 hours in CRCM30 sulfate-free medium (Sigma Chemical Co, St Louis, MO) supplemented with 150 μCi/mL 35S-Na2SO4 (DuPont/NEN, Boston, MA). Mesenteric lymph nodes from three mice (∼60 mg of tissue) were minced and radiolabeled in parallel to KG1a for 8 hours in CRCM30 sulfate-free medium supplemented with 1 mCi/mL 35S-Na2SO4. In additional experiments, KG1a were metabolically radiolabeled for 8 hours with 35S-methionine/cysteine using 150 μCi/mL 35S-methionine/cysteine (DuPont/NEN) in methionine/cysteine-free RPMI 1640 medium (GIBCO-BRL) containing 10% dialyzed FBS. KG1a and murine lymph nodes were lysed in 2% Triton X-100 in Tris-buffered saline (TBS) containing 1 mmol/L phenylmethylsulfonyl fluoride and 1 μg/mL each of aprotinin, leupeptin, and pepstatin A overnight at 4°C. The lysates were clarified by centrifugation at 10,000g for 15 minutes and the samples were precleared with protein G-agarose (GIBCO-BRL) for 4 hours. Immunoprecipitation was performed using equivalent amounts of trichloroacetic acid (TCA)-precipitable counts from lymph node and KG1a lysates, using 2 μg IgM control antibody or MECA 79 followed by 6 μg goat antirat IgM secondary antibody. CD43 was immunoprecipitated with 1 μg anti-CD43 or IgG1 isotype control. Immunoprecipitates were collected by incubation with protein G-agarose overnight at 4°C. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed under reduced conditions followed by autoradiography as previously described.12
Lymphocyte adherence assay.The lymphocyte adherence assay provides a semiquantitative assessment of L-selectin ligand activity and has been described in detail previously.23 Briefly, lymphocyte suspensions (107/mL rat thoracic duct lymphocytes in RPMI 1640 medium) were overlaid onto glass slides containing cytospin preparations of KG1a cells or 8-μm–thick frozen sections of murine lymph node (lymph nodes served as a control to assess the expected L-selectin–mediated lymphocyte binding to HEV). Slides were placed on a rotating platform for incubation under shear (80 rpm) at 4°C for 30 minutes. Slides were then rinsed in PBS to remove nonadherent lymphocytes, fixed in 3% glutaraldehyde, and stained with methyl green-thionin. Slides were examined for lymphocyte adherence to KG1a or lymph node sections by light microscopy. L-selectin ligand activity was measured by counting the number of lymphocytes adherent to a confluent area of KG1a, using an ocular grid under 250× magnification. For measurement of KG1a ligand activity after experimental interventions, untreated (control) cells and treated cells underwent adherence assays in parallel; ligand activity of treated KG1a cells, expressed as the percentage of lymphocyte binding compared to control cells, was assessed by counting respective adherent lymphocytes on several grid areas of KG1a cytospins (minimum of two slides per experiment; three separate experiments). For adherence assays to assess blocking activity of MECA 79, KG1a cytospins or murine lymph node sections were preincubated in RPMI 1640 containing either MECA 79 or isotype control antibody at concentrations as high as 50 μg/mL for 30 minutes at 4°C. Slides were then rinsed and lymphocyte suspensions containing the respective antibodies at concentrations equivalent to that used in the preincubation were overlaid onto the slides.
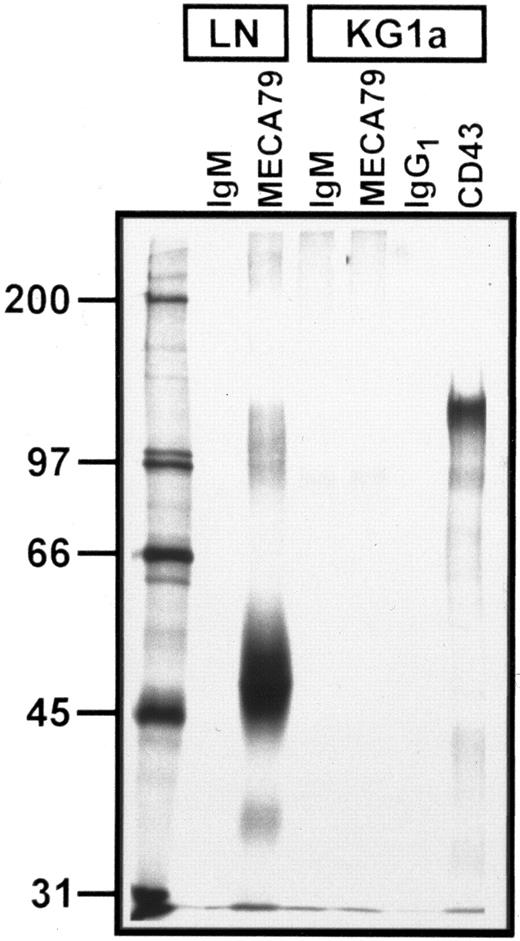
Autoradiograph of SDS-PAGE analysis of immunoprecipitated 35S-SO4 metabolically radiolabeled MECA 79 and CD43 proteins from lymph node and KG1a lysates. Immunoprecipitation with MECA 79, anti-CD43, and isotype control MoAbs are as noted above each lane. Numbers at left are MW markers (in kilodaltons). All immunoprecipitates were normalized for equivalent TCA-precipitable counts. A faint band at ∼90 kD is visualized in both IgM control and MECA 79 immunoprecipitates of KG1a lysates. Note the absence of MECA 79-specific protein in KG1a lysates, but prominent CD43 protein signal.
Autoradiograph of SDS-PAGE analysis of immunoprecipitated 35S-SO4 metabolically radiolabeled MECA 79 and CD43 proteins from lymph node and KG1a lysates. Immunoprecipitation with MECA 79, anti-CD43, and isotype control MoAbs are as noted above each lane. Numbers at left are MW markers (in kilodaltons). All immunoprecipitates were normalized for equivalent TCA-precipitable counts. A faint band at ∼90 kD is visualized in both IgM control and MECA 79 immunoprecipitates of KG1a lysates. Note the absence of MECA 79-specific protein in KG1a lysates, but prominent CD43 protein signal.
Analysis of ligand re-expression after neuraminidase treatment of KG1a.KG1a (20 × 106 cells/mL) were treated with 0.1 U/mL neuraminidase (Vibriocholerae Neuraminidase, 1 U/mL stock; Boehringer Mannheim, Indianapolis, IN) for 1 hour in RPMI 1640 without bicarbonate (GIBCO/BRL; pH 7.4) for 60 minutes at 37°C; for controls, an equivalent volume of enzyme buffer (1/10 vol of 50 mmol/L sodium acetate, 154 mmol/L NaCl, 9 mmol/L CaCl2 , pH 5.5) was added to KG1a in RPMI 1640 without bicarbonate (pH 7.4) under identical conditions (final pH of solution was 6.8). After neuraminidase digestion, cells were washed three times in RPMI 1640 without bicarbonate and an aliquot, designated as t = 0, was tested in the adherence assay to verify complete loss of ligand activity. The remaining cells were cultured in RPMI 1640 with 10% FBS and an aliquot of these cells was removed at serial time points over 24 hours for analysis of the reappearence of ligand activity in the adherence assay.
Lymphocyte Adherence to KG1a
| KG1a Treatment* . | Mean (SEM) of Binding (% of untreated control)† . |
|---|---|
| Neuraminidase (t = 0, no culture) | 0.02 (0.1)‡ |
| Neuraminidase buffer control (no culture) | 98.5 (7.5) |
| Neuraminidase, 24-hr culture | 101.3 (6.5) |
| Neuraminidase, 24-hr culture with chlorate (10 mmol/L) | 107.1 (6.2) |
| Neuraminidase, 20-hr culture with tunicamycin (15 μg/mL) | 0.4 (0.1)‡ |
| Neuraminidase, 20-hr culture with tunicamycin buffer alone | 105.2 (7.5) |
| 20-hr culture with tunicamycin (15 μg/mL) (no neuraminidase pretreatment) | 106.2 (7.2) |
| Neuraminidase, 20-hr culture with cycloheximide (1.25 μg/mL) | 0.5 (0.2)‡ |
| 20-hr culture with cycloheximide (1.25 μg/mL) (no neuraminidase pretreatment) | 91.8 (5.8) |
| KG1a Treatment* . | Mean (SEM) of Binding (% of untreated control)† . |
|---|---|
| Neuraminidase (t = 0, no culture) | 0.02 (0.1)‡ |
| Neuraminidase buffer control (no culture) | 98.5 (7.5) |
| Neuraminidase, 24-hr culture | 101.3 (6.5) |
| Neuraminidase, 24-hr culture with chlorate (10 mmol/L) | 107.1 (6.2) |
| Neuraminidase, 20-hr culture with tunicamycin (15 μg/mL) | 0.4 (0.1)‡ |
| Neuraminidase, 20-hr culture with tunicamycin buffer alone | 105.2 (7.5) |
| 20-hr culture with tunicamycin (15 μg/mL) (no neuraminidase pretreatment) | 106.2 (7.2) |
| Neuraminidase, 20-hr culture with cycloheximide (1.25 μg/mL) | 0.5 (0.2)‡ |
| 20-hr culture with cycloheximide (1.25 μg/mL) (no neuraminidase pretreatment) | 91.8 (5.8) |
After neuraminidase treatment, all cells were washed before culturing as indicated. Experimental details are described in the text.
Number of lymphocytes adherent to confluent area of KG1a were counted by light microscopy using an ocular grid under 250× magnification (quantified a minimum of 2 fields/slide, 2 slides/experiment, 3 separate experiments). Results are presented as the percentage of binding compared with corresponding untreated control KG1a cytospin preparations.
Statistically significant ( P < .0001, by paired t-test) difference compared with neuraminidase buffer control cells. All other values are not statistically different (P > .05).
For studies on the effects of metabolic inhibition of N-linked glycosylation, tunicamycin (Sigma Chemical Co; prepared as 1 mg/mL stock in 95% ethanol) was added to cell cultures (15 μg/mL final concentration, 107 cells/mL in RPMI 1640 with 10% FBS) after either neuraminidase treatment or respective enzyme buffer treatment of the KG1a cells as described above; as control for the effects of ethanol, parallel cultures were established in the presence of 1.5% ethanol. For studies on the effect of protein synthesis inhibition, cycloheximide (Sigma; endotoxin-cleared26 ) was added to cultures of neuraminidase-treated or enzyme buffer-treated KG1a cells at 1.25 μg/mL final concentration and 107 cells/mL in RPMI 1640 with 10% FBS. In each case, cells were cultured for 20 hours and the expression of L-selectin ligand activity was measured by adherence assay at the end of the culture period.
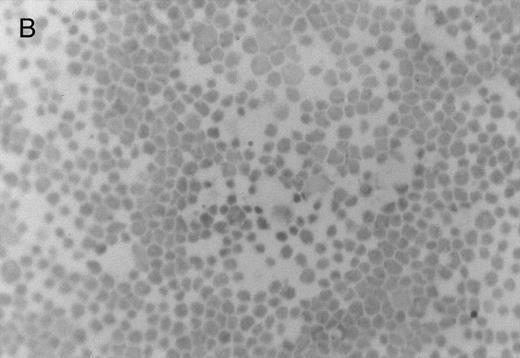
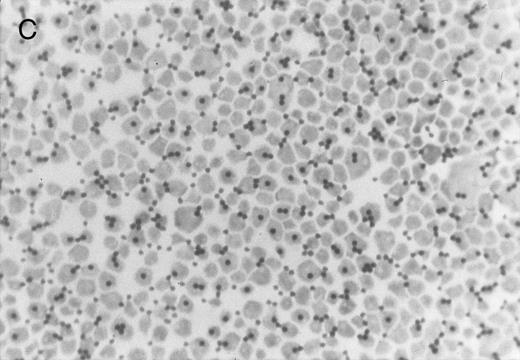
Chlorate inhibition of sulfation does not block KG1a L-selectin ligand activity. Representative results of adherence assay are shown as performed on (A) untreated KG1a; (B) KG1a immediately after neuraminidase treatment; and (C) KG1a 24 hours after neuraminidase treatment, cultured in the presence of 10 mmol/L sodium chlorate. The small solid circles are lymphocytes adherent to large, pale KG1a cells (methyl green-thionine stain, original magnification × 250).
Chlorate inhibition of sulfation does not block KG1a L-selectin ligand activity. Representative results of adherence assay are shown as performed on (A) untreated KG1a; (B) KG1a immediately after neuraminidase treatment; and (C) KG1a 24 hours after neuraminidase treatment, cultured in the presence of 10 mmol/L sodium chlorate. The small solid circles are lymphocytes adherent to large, pale KG1a cells (methyl green-thionine stain, original magnification × 250).
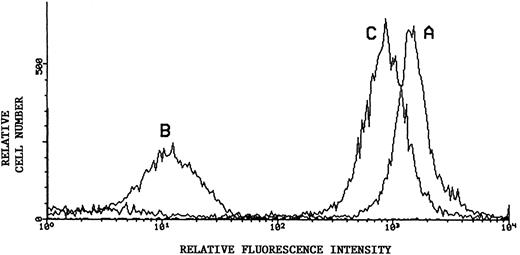
Flow cytometric analysis of sialic acid-dependent epitope of CD43. Representative results using monoclonal L60 as primary antibody in indirect immunofluorescence of KG1a: Untreated KG1a (histogram curve A); KG1a immediately after neuraminidase treatment (histogram curve B); and KG1a 24 hours after neuraminidase treatment, cultured in the presence of 10 mmol/L sodium chlorate (histogram curve C). Secondary antibody was PE-conjugated. Recovery of sialated surface CD43 is evident despite inhibition of sulfation.
Flow cytometric analysis of sialic acid-dependent epitope of CD43. Representative results using monoclonal L60 as primary antibody in indirect immunofluorescence of KG1a: Untreated KG1a (histogram curve A); KG1a immediately after neuraminidase treatment (histogram curve B); and KG1a 24 hours after neuraminidase treatment, cultured in the presence of 10 mmol/L sodium chlorate (histogram curve C). Secondary antibody was PE-conjugated. Recovery of sialated surface CD43 is evident despite inhibition of sulfation.
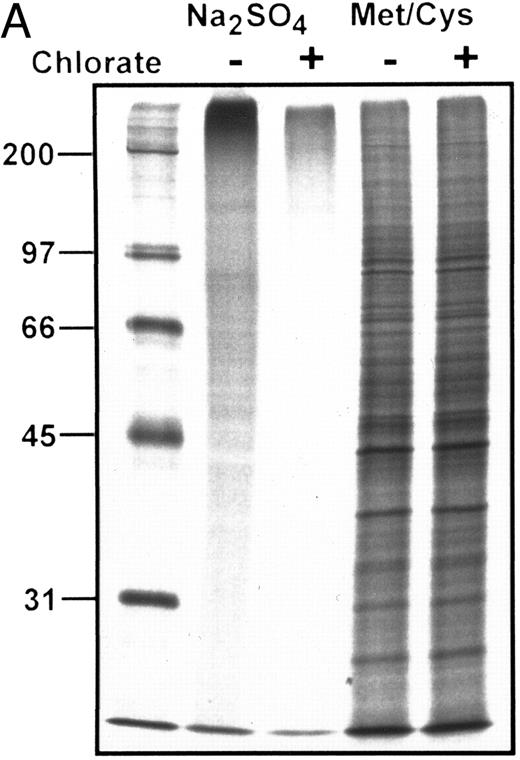
(A) Autoradiograph of SDS-PAGE analysis of total 35S-SO4–radiolabeled and 35S-methionine/cysteine-radiolabeled protein in the presence (+) or absence (−) of chlorate. Lanes contain equivalent amounts of lysate material obtained from cells cultured at identical density in the presence of respective radiolabels with or without chlorate for the terminal 8 hours of the 24 hours of culture (see text for details). Marked inhibition of sulfation is shown among all sulfated proteins in the presence of chlorate, without significant effects on protein synthesis as shown by equivalent profiles of 35S-methionine/cysteine-radiolabeled proteins. MW markers (in kilodaltons) are shown at the left of the figure. (B) Autoradiograph of SDS-PAGE analysis of 35S-SO4 metabolically radiolabeled CD43 in presence or absence of chlorate. CD43 was immunoprecipitated from lysates representing equivalent numbers of chlorate-treated or control (untreated) KG1a cells that were incubated with 35S-SO4 for the terminal 8 hours of the 24 hours of culture. Immunoprecipitations with anti-CD43 or isotype control MoAbs are as noted above each lane; numbers at left are MW markers in kilodaltons. Markedly diminished levels of 35SO4-labeled CD43 are evident in the chlorate-treated cell population.
(A) Autoradiograph of SDS-PAGE analysis of total 35S-SO4–radiolabeled and 35S-methionine/cysteine-radiolabeled protein in the presence (+) or absence (−) of chlorate. Lanes contain equivalent amounts of lysate material obtained from cells cultured at identical density in the presence of respective radiolabels with or without chlorate for the terminal 8 hours of the 24 hours of culture (see text for details). Marked inhibition of sulfation is shown among all sulfated proteins in the presence of chlorate, without significant effects on protein synthesis as shown by equivalent profiles of 35S-methionine/cysteine-radiolabeled proteins. MW markers (in kilodaltons) are shown at the left of the figure. (B) Autoradiograph of SDS-PAGE analysis of 35S-SO4 metabolically radiolabeled CD43 in presence or absence of chlorate. CD43 was immunoprecipitated from lysates representing equivalent numbers of chlorate-treated or control (untreated) KG1a cells that were incubated with 35S-SO4 for the terminal 8 hours of the 24 hours of culture. Immunoprecipitations with anti-CD43 or isotype control MoAbs are as noted above each lane; numbers at left are MW markers in kilodaltons. Markedly diminished levels of 35SO4-labeled CD43 are evident in the chlorate-treated cell population.
Inhibition of sulfation by chlorate.KG1a were treated with neuraminidase as described above and were cultured at 2 × 106 cells/mL in RPMI 1640 containing 10% FBS in the presence or absence of 10 mmol/L sodium chlorate (Sigma Chemical Co) for 24 hours. Adherence assays were performed on chlorate-treated and untreated (control) cells after 24 hours of culture. To verify that lymphocyte binding to chlorate-treated KG1a cells was L-selectin–mediated, adherence assays were performed exactly as described in Oxley and Sackstein23 in the presence of EDTA, of anti–L-selectin blocking MoAbs, of soluble carbohydrate ligands (PPME, fucoidin) to Lselectin, and after PMA-induced shedding of L-selectin off the lymphocyte surface.
To monitor sulfate incorporation in chlorate-treated cells, aliquots of cells from chlorate-incubated and control groups were removed at 8-hour intervals of culture and resuspended in CRCM30 sulfate-free medium containing 150 μCi/mL 35S-Na2S04 with or without chlorate, respectively; metabolic 35SO4 incorporation was measured by TCA precipitation of radiolabeled protein followed by scintillation counting and by SDS-PAGE analysis of aliquots of total lysate material. For the terminal 8-hour time period (16 to 24 hours of culture after neuraminidase treatment), cells incubated with 35S-Na2S04 in the presence or absence of chlorate were lysed for immunoprecipitation of CD43 and SDS-PAGE analysis of 35SO4-labeled CD43 as described above, except that immunoprecipitations from respective control and chlorate-incubated suspensions were normalized for equivalent cell numbers (not TCA counts).
RESULTS
The KG1a L-selectin ligand does not contain MECA 79 determinants.In functional studies, incubation of KG1a cytospin layers with MECA 79 or isotype-control (IgM) rat monoclonal did not inhibit lymphocyte attachment in the adherence assay (eg, for studies using MECA 79 at a concentration of 50 μg/mL, binding was 98.2% ± 3.3% [mean ± SEM] that of control assays without antibody addition). However, as is characteristic of MECA 79,13 lymphocyte attachment to murine lymph node HEV was blocked using MECA 79 at concentrations as low as 2 μg/mL (binding was 15.7% ± 2.8% that of control). To investigate surface expression of MECA 79 antigen on KG1a cells, indirect immunofluorescence staining of cytospin preparations of KG1a was performed and compared with immunofluorescence staining of murine lymph node sections; although lymph node HEV were clearly and characteristically identified by staining with MECA 79, there was no staining evident on KG1a cells (data not shown). In addition, there was no MECA 79 staining evident by flow cytometric analysis of KG1a cells (Fig 1).
To further evaluate for expression of MECA 79 antigens by KG1a, the cells were incubated with either 35S-Na2SO4 or 35S-methionine/cysteine and resultant radiolabeled protein was immunoprecipitated using MECA 79 antibody. As a positive control, murine lymph nodes were radiolabeled, lysed, and immunoprecipitated in parallel with KG1a. To adjust for potential differences in efficiency of radioactive label incorporation between KG1a and murine lymph nodes, the relative amounts of lysate immunoprecipitated from each source were equalized for TCA-precipitable counts. A representative immunoprecipitation of MECA 79 from 35S-Na2SO4–labeled KG1a and lymph node is shown in Fig 2. The characteristic 50,000 and 90,000 MW bands are immunoprecipitated by MECA 79 in lymph node lysate but no MECA 79 bands are detected in the KG1a lysate; similar results were obtained with MECA 79 immunoprecipitation of 35S-methionine/cysteine–labeled KG1a lysates (data not shown). In the figure, the faint band at ∼90,000 MW shown on both MECA 79 and rat IgM-control antibody immunoprecipitates of KG1a cells is nonspecific, because it can be removed by extensive preclearing of lysates with protein-G agarose alone (data not shown); a similar faint 90,000 MW band could also be visualized on IgG1 control immunoprecipitation of 35SO4-labeled KG1a cells. Of note, the sulfated glycoprotein CD43 (∼110 kD) was readily immunoprecipitated from KG1a lysates, verifying that 35SO4 was incorporated into newly synthesized products and that immunoprecipitation conditions were appropriate for the detection of specific radiolabeled KG1a proteins.
The KG1a L-selectin ligand exhibits sulfate-independent binding activity.We have shown previously that the KG1a L-selectin ligand, like the endothelial ligands, requires sialylation for function, because treatment of KG1a with neuraminidase completely abolishes L-selectin–mediated lymphocyte binding.23 We therefore measured the kinetics of recovery of ligand activity after neuraminidase treatment and chose experimental conditions such to maintain chlorate inhibition of sulfation throughout the period when the KG1a ligand is being re-expressed on the cell membrane. Cell viability after neuraminidase digestion was typically greater than 99% by trypan exclusion. Initial studies of the kinetics of recovery of binding activity after neuraminidase treatment showed an absence of ligand activity for 10 to 12 hours; thereafter, ligand activity increased steadily with return to baseline levels within 24 hours. The return of binding activity after desialylation was blocked by metabolic inhibition of N-linked glycosylation by tunicamycin and by inhibition of protein synthesis by cycloheximide (Table 1). The results of tunicamycin and cycloheximide experiments indicate that the membrane turnover of the L-selectin ligand is relatively slow in resting cells, because culture of KG1a in the presence of these reagents for up to 20 hours did not alter L-selectin ligand activity (Table 1). However, after neuraminidase treatment of KG1a, both reagents inhibited return of ligand activity without affecting cell viability (trypan blue exclusion >98%), suggesting that re-expression of sialylated functional ligand after neuraminidase digestion results from de novo synthesis and posttranslational processing of ligand and not from transport of preformed ligand from intracellular compartments to the membrane surface.
The cleavage of sensitive sialic acid epitopes after neuraminidase treatment of KG1a was confirmed by testing for loss of ligand activity in the adherence assay and by measuring the expression of the sialic acid-dependent L60 epitope of CD43 by flow cytometry (Figs 3 and 4 and Table 1). These neuraminidase-treated KG1a were then incubated without chlorate (control) or in presence of chlorate for 24 hours to inhibit sulfation throughout the recovery period for the re-expression of functional L-selectin ligand. Cell viability was greater than 95% in both chlorate-treated and control cell populations (trypan blue exclusion). As shown in Fig 3, 24 hours of chlorate treatment did not alter the re-expression of ligand activity; indeed, quantitation of adherence assays performed on KG1a 24 hours after neuraminidase treatment showed a slightly higher level (although not a statistically significant difference) of L-selectin ligand activity in chlorate-incubated KG1a cells compared with that of nonincubated control cells (Table 1). As with control KG1a cells,23 lymphocyte adherence to chlorate-treated KG1a was L-selectin-specific; it was Ca2+-dependent, was completely inhibited by anti–L-selectin MoAbs known to block L-selectin adhesive function and by carbohydrate molecules known to bind L-selectin (eg, PPME, fucoidin), and was eliminated by phorbol myristate acetate treatment of lymphocytes to induce L-selectin shedding (data not shown; experimental procedures were performed as described in Oxley and Sackstein23 ).
Sequential 35S-SO4 pulse radiolabeling studies at 8-hour time intervals within the 24 hours of chlorate incubation indicated that sulfation was inhibited throughout the entire incubation period, as shown by diminished quantities of TCA-precipitable radiolabeled protein counts (chlorate-incubated counts consistently <10% that of non–chlorate-treated controls) and of total 35SO4-radiolabeled proteins observed by SDS-PAGE/autoradiography of cell lysates (Fig 5A). However, chlorate did not inhibit total protein synthesis, because 35S-methionine/cysteine–incorporated TCA-precipitable counts and autoradiographic profiles of radiolabeled proteins were not significantly different in chlorate and control groups (Fig 5A). As shown in Fig 5B, 35S-SO4 radiolabeling of CD43 was markedly diminished by chlorate treatment, even in the terminal 8 hours of the 24 hours of incubation. However, similar to the results of recovery of ligand activity, chlorate treatment did not block re-expression of CD43 to baseline levels (Fig 4), despite evident sulfate deficiency (Fig 5B). Monitoring of membrane CD43 recovery was performed using a sialylated epitope of the protein, indicating that sialylation of CD43 (like sialylation of the L-selectin ligand) was not affected by chlorate treatment.
DISCUSSION
Previous studies have emphasized the importance of sulfation in the activity of L-selectin ligands, and these results have been underscored by the finding that recognition of functional epitope(s) by MECA 79 antibody is sulfation-dependent.16 Moreover, sulfation is characteristic of both naturally occurring and synthetic molecules that possess L-selectin binding activity, including glycolipids,27 oligosaccharides,28 and glycosaminoglycans,29 and the degree of sulfation correlates with binding affinity.29 In this report, we present evidence that the binding activity of a membrane glycoprotein L-selectin ligand expressed on KG1a cells is sulfation-independent. These data, together with our previous data showing that O-sialoglycoprotease digestion does not affect the KG1a ligand activity,23 defines this glycoprotein distinctly from heretofore characterized L-selectin ligands of endothelial cells and extends our understanding of the structural determinants required for L-selectin recognition/interaction by membrane L-selectin ligands.
The results of our studies provide direct evidence that KG1a cells do not express MECA 79 antigens. Incubation of KG1a with MECA 79 did not interfere with lymphocyte adherence in the binding assay, and immunofluorescence studies showed no evidence of MECA 79 antigen on KG1a. Moreover, no MECA 79-immunoprecipitable proteins were detected from lysates of metabolically radiolabeled KG1a cells using either 35SO4 or 35S-methionine/cysteine, although characteristic bands were obtained from respectively radiolabeled murine lymph nodes. The absence of MECA 79-precipitable protein within KG1a lysates shown here indicates that there are no intracellular accumulations of MECA 79 antigen and, furthermore, that the MECA 79 antigen is not synthesized to any signficant extent by KG1a cells.
Although MECA 79 recognizes a sulfation-dependent epitope, the absence of MECA 79 reactivity does not in itself indicate that sulfation does not participate in the binding determinant of the KG1a ligand. This issue was examined directly by measuring ligand activity after incubation with chlorate, a highly efficient metabolic inhibitor of both saccharide and peptide sulfation.30 Incubation of KG1a in chlorate-containing media was performed over a 24-hour period after complete desialylation of the cells by neuraminidase treatment; because ligand activity is sialic acid-dependent, the recovery of activity after neuraminidase treatment thus reflects de novo ligand expression on the cell surface. The data show that binding activity of the KG1a ligand was unaffected by chlorate incubation. The inhibition of sulfation in KG1a was evident, even for the terminal 8 hours of the incubation period, as shown by a marked reduction in TCA-precipitable 35SO4 counts and by a dramatic decrease in 35SO4 label incorporation among all sulfated proteins as shown by SDS-PAGE analysis (Fig 5A). Despite blockade of posttranslational sulfation by chlorate, it is possible that membrane expression of sulfated KG1a ligand could occur due to transport of preformed sulfated ligand stored within an intracellular compartment; however, this is doubtful because membrane desialylation followed by metabolic inhibition of glycosylation with tunicamycin or of protein synthesis with cycloheximide in each case prevented any re-expression of ligand activity (Table 1). Thus, the re-expression of ligand activity after neuraminidase treatment results predominantly from synthesis/processing of nascent ligand, and the absence of a chlorate effect indicates that sulfation does not contribute significantly to L-selectin recognition by the KG1a ligand. Of note, the profound chorate-induced decrease in 35SO4 incorporation into CD43 (a sialylated and sulfated glycoprotein expressed on KG1a23,31), without disruption in the return to baseline levels of sialylated membrane CD43 in the presence of chlorate (Fig 4), suggests that sialylation and membrane transport of this glycoprotein also occur independently of sulfation. Thus, although our data do not provide information on whether the KG1a L-selectin ligand contains sulfate modifications, the results strongly indicate that, similarly to CD43, the role of any sulfation is not central to the appropriate processing of the L-selectin binding determinant (which includes sialylation) and of membrane transport of de novo synthesized ligand.
The data reported here provide first evidence of a functional membrane glycoprotein L-selectin ligand whose binding activity is not sulfate-dependent. Multiple studies have emphasized the importance of sulfation in the binding determinant of ligands for both L-selectin16,21,27,28,29,32,33 and P-selectin,34-36 and all heretofore identified naturally occurring membrane ligands for these selectins bear sulfate-dependent activity. Although there is evidence that sulfation is critical within the sugar determinants of the endothelial L-selectin ligands,33 it has not been formally shown whether protein sulfation is also of importance in the function of these molecules. However, protein sulfation (at tyrosines) is fundamental in P-selectin ligand activity35-37 and in some cases of integrin-mediated adhesion.38 The contribution of the sulfate modifications may relate to electrostatic forces via the localization of negative ions within discrete molecular determinants. Such a role for charge in L-selectin ligands is supported by the finding that unsulfated, but anionic polysaccharides such as polymers of phosphated mannose can bind to L-selectin.39 Within the KG1a ligand, it is possible that glycosidic and/or amino acid modifications such as phosphorylation or the molecular composition of the discrete sugars or amino acids comprising the binding domain create a relevant anionic milieu. Present efforts are directed at isolating and characterizing the structure of this molecule. Although the precise structural features that direct binding activity for this and other naturally expressed membrane L-selectin ligands remain to be determined, the data presented here demonstrate that determinants conferring high-affinity recognition of L-selectin may vary among different cell types that express such ligands.
ACKNOWLEDGMENT
This report is dedicated to the memory of Dr Yee Hon Chin.
Supported in part by a grant from the American Cancer Society, Florida Chapter.
Address reprint requests to Robert Sackstein, MD, PhD, Division of Bone Marrow Transplantation, H. Lee Moffitt Cancer Center & Research Institute, 12902 Magnolia Dr, Tampa, FL 33612.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal