Abstract
Paroxysmal nocturnal hemoglobinuria (PNH) is characterized by clonal blood cells that are deficient in the surface expression of glycosylphosphatidylinositol-anchored proteins due to somatic mutation in the X-linked gene PIG-A. In some patients, more than one abnormal clone may be present. Analysis of bulk DNA/RNA from granulocytes has been useful in identifying the predominant PIG-A mutation in each patient. However, it is often not useful in determining the presence of minor clones. Many patients have cells with partial deficiency. Here, we analyzed the PIG-A gene in two B-cell lines bearing complete or partial deficiencies, cells of hematopoietic progenitor colonies and peripheral blood granulocytes from the same patient. We found that two B-cell lines had different mutations, the granulocytes contained at least two mutants, and the hematopoietic progenitors contained four mutants. Three of the four were shared by B cells and/or granulocytes whereas the other one was found only in the hematopoietic progenitors. The partial deficiency was caused by a point mutation near an alternative splice site within exon 2 that resulted in partial decreases of activity and quantity of the full-length transcript. These results further show the oligoclonal nature of PNH and differences in extent of expansion among mutant clones.
PAROXYSMAL NOCTURNAL hemoglobinuria (PNH) is an acquired clonal hematologic disorder characterized by intravascular hemolytic anemia.1-3 Abnormal blood cells are deficient in the surface expression of glycosylphosphatidylinositol (GPI)-anchored proteins.4,5 Deficiencies of GPI-anchored complement regulatory proteins, such as decay accelerating factor (DAF ) and CD59, render red blood cells very sensitive to complement and cause complement-mediated hemolysis and hemoglobinuria.3
The X-linked gene PIG-A that is involved in biosynthesis of the GPI-anchor6 is mutated in the affected cells.7 Granulocytes and lymphocytes have the same mutation, indicating that a somatic mutation occurs in a hematopoietic stem cell.7 The PIG-A gene is mutated in all patients with PNH examined to date at the molecular level.8-17 The most likely explanation as to why PIG-A is always responsible is that the PIG-A gene is localized on the X-chromosome,7 therefore, a single loss-of-function mutation in PIG-A would cause a GPI-anchor deficiency because even in a female hematopoietic stem cell, only one X-chromosome is functional due to the somatic inactivation of the X-chromosome. In addition, PIG-A may be the only X-linked gene among 10 or so genes involved in GPI biosynthesis. In fact, four other genes, PIG-H,18 PIG-F19, PIG-B,20 and PIG-C21 have been proven to be autosomal.20-23 Mutations on both alleles of autosomal genes must occur to cause GPI-anchor deficiency, but this event would be very rare.
In the peripheral blood (PB) of patients with PNH, PIG-A mutant cells dominate over normal cells. Earlier studies showed one each of PIG-A mutant clone in most patients.8,9,12 More recent studies showed two or more independent PIG-A mutant clones in some 20% of patients10,13,16 24 (our unpublished observation, June 1996). Although two mutants are similarly dominant in few patients, usually one of the clones predominates. This raises a possibility that minor clone(s) might have been overlooked in many patients.
Some patients with PNH have erythrocytes with partial deficiency of DAF and CD59, termed type II PNH cells. Some of them also have granulocytes and/or lymphocytes that are partially deficient in the surface expression of GPI-anchored proteins. Mutations of the PIG-A gene were also shown in those partially deficient cells.11,12 These mutations are usually point mutations that presumably caused a partial loss of PIG-A activity. Partially deficient erythrocytes exist in 20% to 25% of patients, usually together with completely deficient type III erythrocytes.25 26 If indeed partially and completely deficient erythrocytes coexisting in the same patients are separate mutants of PIG-A, then the majority of patients might have more than one mutant clone.
To get insight into the various extents of dominancy of multiple mutant hematopoietic stem cells in PNH, we have analyzed PIG-A mutations in two B-cell lines bearing a complete and a partial deficiency of GPI-anchored proteins, as well as in cells of hematopoietic progenitor colonies and PB granulocytes all from one patient with PNH. We have shown that this patient has 4 PIG-A mutant clones, 1 predominant and 3 minor clones, 1 of the minor ones being found only in bone marrow (BM) cells.
MATERIALS AND METHODS
Blood cell samples and cell lines.The Epstein-Barr virus–transformed B-lymphoblastoid cell lines TK-4+, -1−, and -14− established from patient J19 with PNH were described previously.27 The human B-lymphoblastoid cell line JY5, a GPI anchor-deficient mutant, was also used.28 Blood samples were obtained from patient J19 after obtaining his informed consent. Granulocytes were isolated by sedimentation through 6% dextran and centrifugation on Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). Forty percent of the granulocytes was completely CD59-deficient. BM mononuclear cell suspensions were also isolated by centrifugation on Ficoll-Hypaque.
Culture of hematopoietic progenitor cells.BM cells (1 × 105) were cultured in methylcellulose to generate burst-forming unit erythroid (BFU-E) bursts, as well as colony-forming unit–granulocyte-macrophage (CFU-GM) and CFU-mix colonies as described.29 Briefly, the cells were plated in 35 mm culture dishes (Flow Laboratory, Rockville, MD) in a 1 mL mixture containing 0.9% methylcellulose (Shinetsu Chemicals Co, Osaka, Japan), 1% deionized bovine serum albumin (Calbiochem, San Diego, CA), 30% fetal calf serum (Microbiological Associates, Bethesda, MD), 0.1 mmol/L 2-mercaptoethanol (Merck, Schuchard, Germany), as well as the recombinant human hemopoietic growth factors, erythropoietin (2 U/mL; Chugai Pharmaceutical Co, Tokyo, Japan), granulocyte colony-stimulating factor (10 ng/mL; Kirin-Amgen, Tokyo, Japan), granulocyte-macrophage colony-stimulating factor (10 ng/mL; Genzyme, Cambridge, MA), interleukin-3 (10 ng/mL; Genzyme) and stem cell factor (10 ng/mL; Genzyme). The culture dishes were incubated at 37°C under 5% CO2 and high humidity. The numbers of colonies/bursts were scored on day 10 for CFU-GM, on day 14 for BFU-E, and on day 16 for CFU-mix after culture initiation. These colonies/bursts were lifted using tapered Pasteur pipets, then single-cell suspensions of individual colonies/bursts were prepared and analyzed for CD59 expression and PIG-A mutations.
Flow cytometry of CD59.Cells were stained for CD59 with biotinylated 5H8 monoclonal antibody30 and with phycoerythrin-conjugated streptoavidin (Biomeda, Foster City, CA). The cells were then analyzed using a FACScan (Becton Dickinson, Lincoln Park, NJ).
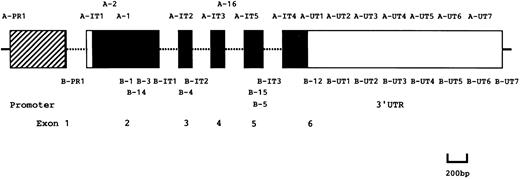
Amplification of the PIG-A gene.DNA was isolated from cell lines (TK-4+, -1−, and -14−), granulocytes and cells from CFU-GM and CFU-mix colonies, as well as BFU-E bursts. Regions of the PIG-A gene were amplified by polymerase chain reaction (PCR) using the primer sets shown in Fig 1 for 35 cycles and the products were used for hetero-duplex and single strand conformation polymorphism (SSCP) analyses or cloned into pBluescript II for sequencing. Sequences of primers were published previously13 31 except those of A-IT5 (5′-TGGTTTTGTTGATCTTCCTG), B-5 (5′-GTTATGGATGTTTTCTGGAGCTG), B-14 (5′-GTAAGCACCGAGCTGACATCAG), and B-15 (5′-TAGCCTTTTCCAATCCTTCA).
Schematic representation of PIG-A gene and PCR primers. ▨, □, and ▪ boxes represent a promoter, untranslated and coding regions, respectively. Primers used for PCR are indicated at the corresponding positions.
Schematic representation of PIG-A gene and PCR primers. ▨, □, and ▪ boxes represent a promoter, untranslated and coding regions, respectively. Primers used for PCR are indicated at the corresponding positions.
Hetero-duplex analysis.The coding regions of PIG-A were amplified by PCR in five fragments using the primer sets, A-IT1 and B-1, A-1 and B-IT1, A-IT2 and B-IT2, A-IT3 and B-IT3, and A-IT4 and B-12 (Fig 1). In some experiments, a region spanning exons 4 and 5 was amplified using the primer set, A-16 and B-5 (Fig 1). For hetero-duplex formation, the PCR products from abnormal cell lines (TK-1− and -14−) were mixed with the corresponding PCR products from the normal cell line (TK-4+) to obtain an approximately 1:1 ratio of the mutant to the normal products, denatured for 3 minutes at 95°C, then slowly cooled to 37°C and kept at that temperature for 1 hour. These samples were resolved by electrophoresis in a Mutation Detection Enhancement gel (MDE; FMC Bioproducts, Rockland, ME) for 4 hours at 800 V, then bands were visualized by staining with ethidium bromide. If a region containing a mutation was detected, the DNA fragment was cloned into pBluescript II for sequencing. The 3′ untranslated region of PIG-A was amplified in a similar manner using the primer sets, A-UT1 and B-UT1, A-UT2 and B-UT2, A-UT3 and B-UT3, A-UT4 and B-UT4, A-UT5 and B-UT5, A-UT6 and B-UT6, and A-UT7 and B-UT7 (Fig 1).
SSCP.Regions spanning exon 2 and exons 4 and 5 of PIG-A, respectively, were amplified by PCR using the nested primer sets. First amplifications proceeded with nonradioactive deoxyribonucleotide triphosphate (dNTP) and primer sets A-2 and B-3, and A-IT3 and B-IT3, and second amplifications were with [α-32P]deoxycytidine triphosphate (dCTP) and the primer sets, A-1 and B-14, and A-IT5 and B-15 (Fig 1), respectively. The PCR products were mixed with an equal volume of a solution containing 95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol. The mixtures were heat-denatured at 95°C, snap-frozen on dry ice and applied to a 6% polyacrylamide gel containing 90 mmol/L Tris-borate buffer (pH 8.3) and 10% glycerol. Electrophoresis proceeded at 3 to 6 W for 10 to 16 hours at 25°C.The gel was dried on filter paper and exposed to Fuji RX 100 X-ray film at −80°C for 12 to 48 hours with an intensifying screen. If a region containing a mutation was detected with PCR products, DNA clones that showed a band shift were cloned into pBluescript II for sequencing.
Transfection.The functional activity of PIG-A cDNA was assayed by cloning it into the Epstein-Barr virus-based mammalian expression vector pEB and transfecting it into the PIG-A–deficient mutant human B-lymphoblastoid cell line JY5.8 13 The cells (5 × 106) were mixed with 10 μg of pEB bearing PIG-A cDNA in 0.8 mL of HeBS transfection buffer (20 mmol/L HEPES, pH 7.05-137 mmol/L NaCl-5 mmol/L KCl-0.7 mmol/L Na2HPO4 -6 mmol/L dextrose). Electroporation was performed at 960 μF and 250 V with a Gene Pulser (Bio-Rad Laboratories, Hercules, CA). After culture and selection with 400 μg/mL hygromycin B for 10 to 14 days, the transfected cells were stained for CD59 to assess complementation of the PIG-A–deficient phenotype of the mutant.
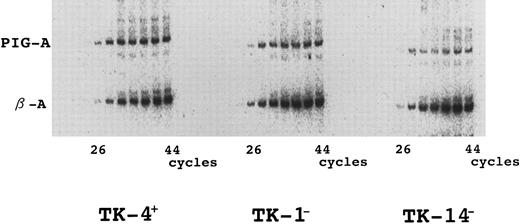
Quantitative reverse-transcriptase PCR (RT-PCR) of the PIG-A mRNA.To quantify the expression of PIG-A mRNA, total RNA was reverse transcribed with random primers (Takara, Kyoto, Japan), and PIG-A and β-actin cDNA were amplified by PCR using [α-32P]dCTP. During the initial 10 cycles of the reactions, in which one cycle consisted of 1 minute of denaturation at 94°C, 1 minute of annealing at 60°C, and 2 minutes of extension at 72°C, only PIG-A cDNA was amplified with primers A-1 and B-4.13 Primers β-A (5′-AAGAGAGGCATCCTCACCCT) and β-B (5′-TACATGGCTGGGGTGTTGAA) for β-actin were added, and PCR was continued for 34 more cycles to amplify both β-actin and PIG-A. Samples removed after every three cycles from 26 to 44 cycles were resolved by polyacrylamide gel electrophoresis and visualized by autoradiography.
Assay of the promoter-regulatory region using a luciferase reporter gene.A 0.66-kb fragment of the promoter region amplified from genomic PIG-A using primers A-PR1 and B-PR131 was cloned into pBluescriptII. The fragment of promoter region was then ligated into a HindIII-and Bgl II-digested luciferase reporter plasmid (PGV-C or PGV-P) (Toyo Ink, Tokyo, Japan). Samples of 1 × 107 JY5 cells were mixed with 20 μg of each PIG-A–promoter luciferase construct, then electroporated at 960 μF and 250 V. After a 48-hour culture, cells were washed twice with phosphate-buffered saline, lysed in 25 mmol/L Tris-phosphate (pH 7.8), 2 mmol/L dithiothreitol, 2 mmol/L 1,2-diamino-cyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol and 1% Triton X-100 (Sigma, St Louis, MO), then centrifuged.31 Luciferase activity in the supernatants was then measured according to the manufacturer's protocol.32
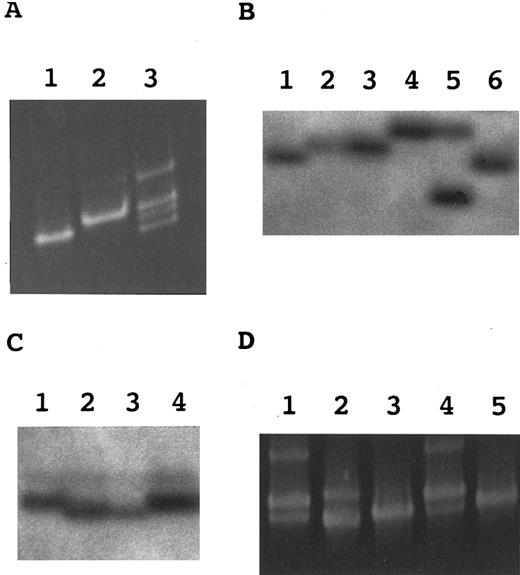
Hetero-duplex and SSCP analyses. (A) Hetero-duplex analysis of a region spanning exons 4 and 5. Lane 1, TK-4+; lane 2, TK-14−; lane 3, a mixture of TK-4+ and TK-14−. (B) SSCP analysis of exon 5 in marrow hematopoietic progenitor cells. Lane 1, TK-4+; lane 2, TK14− (mutation 1); lane 3, a CD59− CFU-mix; lane 4, a CD59− BFU-E; lane 5, another CD59− BFU-E; lane 6, a CD59+ BFU-E. (C) SSCP analysis of exon 2 in marrow hematopoietic progenitor cells. Lane 1, TK-4+; lane 2, TK-1− (mutation 2); lane 3, a CD59− BFU-E; lane 4, a CD59+ BFU-E. (D) Hetero-duplex analysis of a region spanning exons 4 and 5 in the PB granulocytes. Lanes 1 through 3, mixtures of TK-4+ and three clones from granulocytes; lane 4, a mixture of TK-4+ and TK-14−; lane 5, TK-4+.
Hetero-duplex and SSCP analyses. (A) Hetero-duplex analysis of a region spanning exons 4 and 5. Lane 1, TK-4+; lane 2, TK-14−; lane 3, a mixture of TK-4+ and TK-14−. (B) SSCP analysis of exon 5 in marrow hematopoietic progenitor cells. Lane 1, TK-4+; lane 2, TK14− (mutation 1); lane 3, a CD59− CFU-mix; lane 4, a CD59− BFU-E; lane 5, another CD59− BFU-E; lane 6, a CD59+ BFU-E. (C) SSCP analysis of exon 2 in marrow hematopoietic progenitor cells. Lane 1, TK-4+; lane 2, TK-1− (mutation 2); lane 3, a CD59− BFU-E; lane 4, a CD59+ BFU-E. (D) Hetero-duplex analysis of a region spanning exons 4 and 5 in the PB granulocytes. Lanes 1 through 3, mixtures of TK-4+ and three clones from granulocytes; lane 4, a mixture of TK-4+ and TK-14−; lane 5, TK-4+.
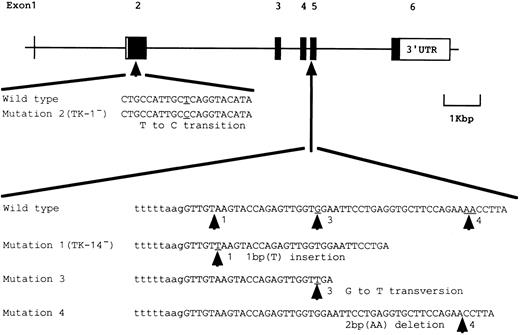
Schematic representation of somatic mutations of the PIG-A gene found in patient J19.
Schematic representation of somatic mutations of the PIG-A gene found in patient J19.
RESULTS
Somatic mutations in two B-cell lines having complete (TK-14−) and partial (TK-1−) deficiencies.We analyzed DNA from TK-14− and TK-1− by a hetero-duplex assay to identify somatic mutations of PIG-A in these phenotypically different B-cell lines. A mixture of DNA segments spanning exons 4 and 5 from TK-14− and a wild-type line TK-4+ formed hetero-duplexes (Fig 2A, lane 3). Sequence analysis showed a T insertion in codon 329 (termed mutation 1) that caused a frameshift and a premature stop codon 25 to 27 nucleotides downstream (Fig 3).
A mutation in TK-1− was not shown by the hetero-duplex analysis, so we sequenced all the coding regions of the PCR-amplified DNA and found a T to C transition in codon 113 (termed mutation 2) that caused a change from leucine to proline (Fig 3).
Somatic mutations of PIG-A in BM hematopoietic progenitor cells.To see whether mutations 1 and 2 found in the B-cell lines are shared by cells in other lineages, we analyzed PIG-A DNA in hematopoietic progenitor cells. Eight bursts of BFU-E, nine colonies of CFU-GM, and eight of CFU-mix were obtained from nonphagocytic mononuclear marrow cells. Single cell suspensions were prepared from these colonies/bursts. Half of each cell suspension was used for flow cytometric analysis of CD59 expression and the other half was used for SSCP analysis. Flow cytometry showed that four of 25 colonies/bursts (one BFU-E and three CFU-GM) were CD59+, wild-type, and that 21 were CD59-deficient (data not shown).
SSCP analysis of PCR-amplified DNA fragments containing exon 5 showed three groups of shifted bands (Fig 2B, lanes 3 through 5). Thirteen of 21 CD59-deficient colonies/bursts (two BFU-E, five CFU-GM, and six CFU-mix) showed the same profile as TK-14− (lanes 2 and 3). Four (two each of BFU-E and CFU-mix) and two (BFU-E) were different (lanes 4 and 5). Sequence analysis of these shifted bands showed mutation 1 in the first, dominant group but not in the other two. Since this mutation was shared by TK-14− B cells and all three types of colonies/bursts, it should have occurred in a hematopoietic stem cell.
Different mutations were found in exon 5 in two other groups of colonies/bursts (Fig 4) with different band shifts (Fig 2B, lanes 4 and 5). The second group had a G to T transversion in codon 335 (Fig 4, panel 1) that generated a stop codon, a nonsense mutation (termed mutation 3; Fig 3). The third group had a two-base (AA) deletion from codon 343 (Fig 4, panel 2) that caused a frameshift and a premature stop codon 13 nucleotides downstream (termed mutation 4; Fig 3).
Mutations 3 and 4 in exon 5 of PIG-A found in marrow hematopoietic progenitor cells. (1) A G to T transversion (mutation 3) indicated by the left arrow; (2) a 2-bp (AA) deletion (mutation 4) indicated by the right arrow.
Mutations 3 and 4 in exon 5 of PIG-A found in marrow hematopoietic progenitor cells. (1) A G to T transversion (mutation 3) indicated by the left arrow; (2) a 2-bp (AA) deletion (mutation 4) indicated by the right arrow.
As shown in Fig 2C, SSCP analysis of exon 2 in 21 CD59-deficient colonies/bursts showed a shifted band in one each of BFU-E (lane 3) and CFU-GM (not shown) which was aligned with that of TK-1− (lane 2), indicating that these two colony/bursts had mutation 2. This mutation was therefore shared by B cells and erythroid and myeloid progenitors, indicating that it was also a mutation in a hematopoietic stem cell.
Somatic mutations of PIG-A in the PB granulocytes.To determine whether these additional mutations 3 and 4 are present in the PB cells, we amplified exon 5 from DNA of granulocytes using primer set A-16 and B-5, cloned the products into pBluescript II and obtained 27 clones. The same region was amplified again from these cloned DNA and analyzed for hetero-duplex formation on mixing with the corresponding normal fragment. There were three profiles as shown in Fig 2D. Fourteen of 27 clones (lane 1) showed the same profile as TK-14− (lane 4) and hence had mutation 1. Two were different (lane 2) and mutation 3 was found in these clones by sequencing. The remaining 11 (lane 3) did not form hetero-duplexes, showing one band as a wild-type control (lane 5). Therefore, mutation 3 was shared by granulocytes, BFU-E and CFU-mix, indicating that it occurred in a myeloid or more primitive stem cell. On the other hand, mutation 4 was found only in BM hematopoietic progenitor cells.
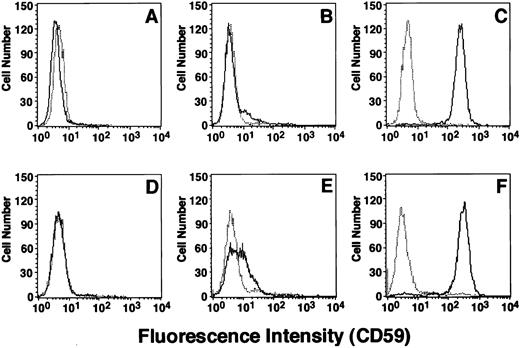
Consequences of mutations 1 and 2; losses of activity and quantity of PIG-A mRNA.Mutations 1 and 2 were found in TK-14− and TK-1− cells, which were completely and partially deficient in the surface expression of GPI-anchored proteins, respectively. To see the effects of these mutations on the activity of PIG-A, we cloned mutant cDNAs into a mammalian expression vector, transfected them into the PIG-A deficient JY5 cells and assessed the restoration of the surface expression of CD59. PIG-A cDNA bearing mutation 1, a T insertion, did not restore the surface CD59 (Fig 5A), whereas wild-type cDNA (C) restored a normal level of expression (F ), indicating that this mutation that caused a premature stop codon resulted in a complete loss of function. A cDNA bearing mutation 2, a leucine to proline substitution, restored a low level of CD59 expression (B), indicating that this amino acid change caused a great decrease of activity. The extent of the activity loss caused by these mutations (A and B), therefore, correlated with the extent of the surface deficiency of GPI-anchored proteins on the patient's cells (D and E).
Flow cytometry of CD59 on JY5 cells transfected with mutant PIG-A cDNA and on B-lymphoblastoid cell lines established from patient J19. (A) The PIG-A–deficient human lymphoblastoid cell line JY5 transfected with PIG-A cDNA bearing mutation 1. (B) The PIG-A–deficient human lymphoblastoid cell line JY5 transfected with PIG-A cDNA bearing mutation 2. (C) JY5 transfected with normal PIG-A cDNA. (D through F ) The completely deficient TK-14−, the partially deficient TK-1− and the wild-type TK-4+–lymphoblastoid cell lines derived from this patient, respectively. Solid lines, anti-CD59; dotted lines, isotype-matched control monoclonal antibody.
Flow cytometry of CD59 on JY5 cells transfected with mutant PIG-A cDNA and on B-lymphoblastoid cell lines established from patient J19. (A) The PIG-A–deficient human lymphoblastoid cell line JY5 transfected with PIG-A cDNA bearing mutation 1. (B) The PIG-A–deficient human lymphoblastoid cell line JY5 transfected with PIG-A cDNA bearing mutation 2. (C) JY5 transfected with normal PIG-A cDNA. (D through F ) The completely deficient TK-14−, the partially deficient TK-1− and the wild-type TK-4+–lymphoblastoid cell lines derived from this patient, respectively. Solid lines, anti-CD59; dotted lines, isotype-matched control monoclonal antibody.
Northern blotting of PIG-A mRNA indicated that TK-14− cells express no detectable PIG-A mRNA and that TK-1− cells express a very low level.7 We performed semiquantitative RT-PCR to confirm these quantitative abnormalities of PIG-A mRNA. These quantitative abnormalities were also seen in this analysis as a severe and a mild decrease of PIG-A mRNA in TK-14− and TK-1− cells, respectively (Fig 6). Quantitative abnormalities could be due either to low levels of transcription or to a shorter life-time of transcripts. Such abnormalities might be caused by a mutation in the promoter or the 3′ untranslated region. To test the former notion, we amplified the promoter region from DNA of TK-14−, TK-1−, and TK-4+ by means of PCR using primer set A-PR1 and B-PR1, cloned them into a reporter luciferase plasmid and performed the luciferase assay. There was no significant difference in their activities, eliminating this possibility (data not shown). We then searched for mutations within the 3′ untranslated region. The 3′ untranslated region was amplified in seven overlapping segments by means of PCR using the primer sets shown in Fig 1, then hetero-duplexes were analyzed. No mutations were indicated in this region in TK-14− and TK-1− cells (data not shown). These results suggested that the coding mutations themselves may also be responsible for the quantitative abnormalities.
Quantitative abnormalities of PIG-A transcripts in TK-1− and TK-14−. RNA was reverse transcribed with random primers and PIG-A and β-actin mRNA were amplified by PCR. During PCR, radiolabeled amplification products were removed after every three cycles, separated by electrophoresis in polyacrylamide gel, and visualized by autoradiography. The bands corresponding to PIG-A and β-actin are indicated. Left panel, TK-4+; center panel, TK-1−; right panel, TK-14−.
Quantitative abnormalities of PIG-A transcripts in TK-1− and TK-14−. RNA was reverse transcribed with random primers and PIG-A and β-actin mRNA were amplified by PCR. During PCR, radiolabeled amplification products were removed after every three cycles, separated by electrophoresis in polyacrylamide gel, and visualized by autoradiography. The bands corresponding to PIG-A and β-actin are indicated. Left panel, TK-4+; center panel, TK-1−; right panel, TK-14−.
DISCUSSION
In this investigation we analyzed PIG-A abnormalities in one patient to address two current issues regarding characteristics of PNH: the extent of oligoclonality and the basis of seemingly frequent partial deficiency. We previously established two phenotypically different B-cell lines from a male patient with PNH, J19.27 TK-14− cells are completely deficient in the surface expression of GPI-anchored proteins and have no detectable PIG-A mRNA as assessed by Northern blotting.7 TK-1− cells are partially deficient, expressing a few percent of normal levels of GPI-anchored proteins, and about 10% of the normal level of PIG-A mRNA.7 First, we examined the number of PIG-A mutant clones in this patient by analyzing these B-cell line, BM hematopoietic progenitor cells, and PB granulocytes and found at least four mutant clones. Second, we found a new mutation that is responsible for partial deficiency.
TK-14− and TK-1− cells had different PIG-A mutations. The mutation of TK-14−, mutation 1, was found in the majority of hematopoietic progenitor cells (13 of 25 colonies/bursts) and of granolucytes (14 of 27 clones) (Table 1). The mutation of TK-1−, mutation 2, was found in two of 25 colonies/bursts (Table 1). Two other clones bearing mutations 3 and 4 were found in four and two of 25 colonies/bursts, respectively (Table 1). Mutations 1 and 2 should have occurred in hematopoietic stem cells because they were shared by B cells and erythroid/myeloid cells. Mutation 3 may have also occurred in hematopoietic or myeloid stem cell because it was shared by CFU-mix, erythroid cells, and granulocytes. Mutation 4 was found only in erythroid bursts, so if it is a mutation in a hematopoietic stem cell remains unclear. Therefore, this patient has four PIG-A mutant clones, at least two of which were hematopoietic stem cell mutants.
Somatic Mutations of PIG-A Found in Patient J19
| Mutation . | Nucleotides . | Nucleotide Change . | Codon . | Consequence . | Frequency of Mutations . | |
|---|---|---|---|---|---|---|
| . | . | . | . | . | Hematopoietic Progenitor Colonies/Bursts . | Granulocyte DNA Clones . |
| Mutation 1 (TK-14−) | 987 T | 1 bp Insertion | 329 | Frame shift | 13/25 | 14/27 |
| Mutation 2 (TK-1−) | 338 T to C | Transition | 113 | Missense (Leu to Pro) | 2/25 | ND |
| Mutation 3 | 1003 G to T | Transversion | 335 | Nonsense | 4/25 | 2/27 |
| Mutation 4 | 1028 AA | 2 bp Deletion | 343 | Frame shift | 2/25 | 0/27 |
| Mutation . | Nucleotides . | Nucleotide Change . | Codon . | Consequence . | Frequency of Mutations . | |
|---|---|---|---|---|---|---|
| . | . | . | . | . | Hematopoietic Progenitor Colonies/Bursts . | Granulocyte DNA Clones . |
| Mutation 1 (TK-14−) | 987 T | 1 bp Insertion | 329 | Frame shift | 13/25 | 14/27 |
| Mutation 2 (TK-1−) | 338 T to C | Transition | 113 | Missense (Leu to Pro) | 2/25 | ND |
| Mutation 3 | 1003 G to T | Transversion | 335 | Nonsense | 4/25 | 2/27 |
| Mutation 4 | 1028 AA | 2 bp Deletion | 343 | Frame shift | 2/25 | 0/27 |
Abbreviation: ND, not done.
Four PIG-A mutant clones had different extents of expansion. A clone with mutation 1 was predominant, contributing to the majority of the hematopoietic progenitor cells and PB granulocytes. Three other mutants were minor. In particular, mutation 4 was found only in the erythroid progenitor cells. It is not known whether erythrocytes derived from this mutant clone were present in the PB. It is possible that the mutant progenitor cells that were in a dormant state in BM produced erythroid bursts under in vitro culture conditions. There is a report that some patients with aplastic anemia who had DAF/CD59-deficient cells in BM but not in the PB eventually developed PNH, ie, showed DAF/CD59-deficient cells in the PB.33 The clone with mutation 4 may eventually become evident in the PB.
In TK-1− cells, we found a substitution of nucleotide 338T to C that caused an amino acid change from leucine to proline. PIG-A cDNA bearing this mutation was weakly active in restoring the surface expression of CD59 on JY5 cells after transfection. This leucine residue is conserved in the mouse PIG-A homologue, Pig-a, and a yeast homologue, GPI3/SPT14 has isoleucine in this position.34 Therefore, this leucine would be important for PIG-A activity and its substitution to proline would account for the partial deficiency. However, this base substitution may also be responsible for the decreased level of PIG-A mRNA since we found no abnormality in the promoter and the 3′ untranslated regions in this cell line. The sequence AGGT at nucleotides 340 to 343 is an alternative splicing site within exon 2.8 9 The base change at nucleotide 338 may have increased splicing at this site with a concomitant decrease of the full-length mRNA. Two characterized patients (HH89 and J1413) had a GC to T change at nucleotides 336/337 and showed a similar skewed profile of splicing. Therefore, it seems likely that some mutations around two alternative splice sites in exon 2 would cause preferential alternative splicing. If such mutations are silent, or affect the activity of PIG-A only slightly, the outcome would be a partial deficiency. This mechanism may account for some cases of the partial deficiency.
We found a single base insertion at nucleotide 987 of PIG-A in TK-14− that caused a frameshift and a premature stop codon. PIG-A cDNA bearing this mutation did not restore the surface expression of CD59 on JY5 cells, indicating that this mutation caused a complete deficiency of TK-14− cells. To understand the basis for the lack of PIG-A mRNA seen in this cell line, we analyzed the promoter region and found that it had normal activity, eliminating the possibility of transcriptional abnormality. Because PIG-A has a long 3′ untranslated region (nearly 3 kb), which may have a role in mRNA stability, we analyzed that region in TK-14− and found no mutation. Thus, we concluded that the base insertion in the coding region may also be responsible for the lack of mRNA and that a premature stop codon may have caused instability of PIG-A mRNA.35,36 A similar mutation was previously found in the British patient HH5.9
We identified four mutant clones in hematopoietic progenitor colonies/bursts. Three of them were also found in B-cell lines and/or granulocytes. Therefore, analysis of PIG-A using hematopoietic progenitor colonies/bursts appeared to be a reliable means with which to identify somatic mutations of PIG-A and to determine the number of mutants in each patient. Analysis of PIG-A using BFU-E bursts should also be useful to determine the somatic mutations responsible for partial deficiency of type II erythrocytes seen in the PB of many patients.
Four independent PIG-A mutants have been found in T-cell clones established from one patient with PNH.24 Thus, the presence of four independent mutant clones does not seem to be exceptional. Taken together with the previous suggestion that a majority of the patients would have more than one PIG-A mutant clone,4 we may describe that PNH is an oligoclonal hematopoietic stem cell disease harboring one to four PIG-A mutants.
The mechanism of clonal expansion in PNH is not well understood. A study with chimeric mice bearing Pig-a–deficient hematopoietic cells suggested that Pig-a mutation alone is not sufficient for clonal expansion.37 The fact that multiple PIG-A mutant clones expand in patients suggests that PIG-A mutation or deficient surface expression of GPI-anchored proteins is relevant to clonal expansion. It was hypothesized that some selection mechanism that allows selective survival of PIG-A mutants may operate in BM of patients with PNH.10 Another which is not mutually exclusive with this mechanism is that mutant clones harbor another genetic change(s) as well as PIG-A mutation that together confer an ability to expand on mutants.38 Further study is necessary to clarify this issue.
Supported by grants from the Ministry of Education, Science, Sports and Culture of Japan.
Address reprint requests to Jun-ichi Nishimura, MD, PhD, Department of Hematology and Oncology, Osaka University Medical School, 2-2 Yamada-oka, Suita, Osaka 565, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal