Abstract
In the biology of a cell, the central role of p53 in controlling functions such as G1/S transition (check point) and DNA damage repair, and as a trigger of apoptosis, is well established. Somatic mutations or other changes in P53 have been reported in numerous tumor types, and in some of these, they are associated with poor prognosis. In this study, we examined 237 cytogenetically characterized B-cell non-Hodgkin's lymphomas (B-NHLs) for somatic changes in P53 by Southern blot analysis, by single-strand conformation polymorphism analysis (SSCP) of exon 5 through 9, and by direct sequencing of SSCP variants to determine the frequency and types of mutations and their clinical significance. In a portion of these (173 tumors), we also studied p53 expression by immunostaining. On Southern blots, no gross change was identified in P53 and no mutation was identified in exon 9. In exons 5 through 8, 27 different mutations were identified in 25 patients (23 single-base substitutions, 3 deletions, 1 duplication). Mutations in P53 were identified in 25 of 237 tumors (10.5%), which included 1 of 45 small lymphocytic lymphomas (SLLs), 2 of 38 follicular small cleaved-cell lymphomas (FSCCs), 2 of 35 follicular mixed small cleaved-cell and large-cell lymphomas (FMxs), 1 of 4 follicular large-cell lymphomas (FLCs), 1 of 14 diffuse small cleaved-cell lymphomas (DSCCs), 2 of 17 diffuse mixed small- and large-cell lymphomas (DMxs), and 16 of 84 diffuse large-cell lymphomas (DLCCs); the difference between the histologic groups was significant (P < .01). Among mantle-cell lymphoma (MC) patients, 3 of 10 had mutations. In 16 patients, the mutation was identified in specimens obtained at diagnosis. Mutation of transition type and transversion type occurred at a relative frequency of 2:1. Thirty percent occurred at CpG dinucleotide sequences and the codon for arginine was most frequently affected. Nineteen of 99 tumors with complex cytogenetic abnormalities, but none of 69 tumors with simple cytogenetic abnormalities, had mutations (P < .001). Similarly, 11 of 25 tumors with an abnormality of 17p and 8 of 143 tumors with apparently normal 17p had mutations (P < .0001). Positive correlations were found between a mutation and p53 expression (P < .001), between missense type mutations and p53 expression (P < .005), and between 17p abnormalities and p53 expression (P < .05). Twenty-two of 49 patients without mutation and 14 of 17 patients with mutations died (P < .05), but there was no significant difference in median survival. Similarly, 21 of 26 p53 positive patients died, whereas only 1 of 24 p53-negative patients died on-study (P < .001). Among p53-negative patients, mutation (P < .01) was positively associated with a fatal outcome. These findings indicate that in B-NHL, somatic changes in P53 were present in diagnostic specimens of all histologic types, but at a higher frequency in DLC and MC tumors. P53 mutation and/or expression has a negative influence on survival, and therefore can serve as prognostic indicators. Immunostaining for p53 is an effective way to screen for P53 changes in these tumors.
SOMATIC GENETIC CHANGES that impair the action of negative growth regulators play critical roles in tumorigenesis. In contrast to dominantly acting proto-oncogenes, which have been implicated in initiation, changes in recessively acting tumor-suppressor genes have been particularly implicated in progression of disease or in development of malignant clones refractory to treatment. In relation to the latter group, P53 has been found to be altered in a wide variety of tumors and in some tumors it has been associated with a poor prognosis.1-3 Besides deletion or rearrangement, which lead to loss of function, dominantly acting negative point mutations in coding regions whose products inhibit the action of the normal allele are well known.4 5
Mutations in P53 with a frequency of 5% (myelodysplastic syndromes) to 50% (acute lymphoblastic leukemia–L3 type) have been reported in various hematologic malignancies.4,5 In the majority of these cases, a mutated P53 has been associated with advanced disease or with the development of resistance to therapy.3 6-9
B-cell non-Hodgkin's lymphoma (B-NHL) occurs in several histologic types ranging from indolent low-grade tumors to highly malignant immunoblastic tumors. Cytogenetic studies performed by us and by others have identified recurring chromosome changes, particularly reciprocal chromosome translocations affecting BCL1, BCL2, BCL3, BCL6, and cMYC, that are frequently associated with specific histologic groups.10,11 The majority of low-grade follicular lymphomas transform into more aggressive histologic types, such as diffuse large-cell lymphoma (DLC), and are then usually associated with a poor prognosis.12-18 Cytogenetic studies have identified secondary nonrandom chromosome changes such as del(6q), del(17p) in some cases, which developed with disease progression.19-23 Although loss of chromosome 17p to which P53 was mapped has been reported in high-grade lymphoma, the status of P53 has not been systematically examined in B-NHL.5,24-26 A few studies have examined gross structural changes27 or mutations in P53 in selected histologic groups of lymphoma.17,18 28-32 Therefore, we examined 237 cytogenetically characterized, but unselected B-NHL tumors for somatic changes in P53 (1) by Southern blot analysis for gross genomic changes; (2) for point mutations in exons 5 through 9 by polymerase chain reaction (PCR) amplification–single-strand conformation polymorphism (SSCP) analysis; (3) by direct sequencing of PCR-amplified genomic DNA from SSCP variants; and (4) for the expression of p53 by immunostaining to characterize the spectrum of somatic genetic changes in this gene.
MATERIALS AND METHODS
Tumor samples. Between January 1989 and December 1994, 237 biopsy samples of lymph node or spleen or other tissues diagnosed histologically as B-NHL as described in the Working Formulation and in the recently published REAL classification33,34 have been studied by cell-surface markers, by genotyping, and by chromosome analysis. Cytogenetic studies were performed on cell suspensions prepared by mincing the fresh tissue in RPMI 1640. Metaphase spreads were obtained by harvesting lymphoma cells directly (without further culturing) or after culturing cells overnight in RPMI 1640 supplemented with 10% fetal calf serum, 1% L-glutamine, and 1% penicillin. Karyotypes were described following ISCN.35
Southern blot analysis. Structure of the P53 gene was analyzed by Southern blot analysis. Eight micrograms to 10 μg of high–molecular-weight DNA prepared by standard procedures was digested with restriction endonucleases, size-fractionated through 0.8% tris-boric acid-EDTA (TBE) gels, denatured, neutralized, and blotted to nitrocellulose. Blots were hybridized with DNA probes labeled with α[32P]dCTP by random priming, washed, and autoradiographed as previously described.36 DNA probes used in these studies were a 5.5-kb BamHI-HindIII fragment covering the J-region of immunoglobulin (Ig) heavy chain,37 a 2.5-kb EcoRI probe for the constant region,38 or a 1.8-kb probe for the J-region (Oncor, Gaithersburg, MD) of Ig kappa chain, 3.7-kb EcoRI-HindIII kb probe for the constant region of Ig lambda gene,39 and a 1,587-bp EcoRI-BamHI probe for P53.40
Oligonucleotide primers. The nucleotide sequence of the primers used in this study has been reported previously.41 Oligonucleotide primers for exons 5 through 9 were synthesized in the core facility of our institution using an Applied Biosystems (Foster City, CA) synthesizer or purchased from GIBCO-BRL (Gaithersburg, MD).
PCR-SSCP. Approximately 100 ng of tumor DNA was amplified by PCR in a final volume of 10 μL using 10 pmol of each primer, 5 μmol/L of dNTPs, 10 mmol/L TrisCl (pH 8.0), 50 mmol/L KCl, 1.0 to 1.5 mmol/L MgCl2 , 0.01% gelatin, 1 μCi of α[32P]dCTP, and 0.05 U of Taq polymerase in a PC-100 programmable thermal cycler (M.J. Research, Watertown, MA) under the following conditions: denaturation for 1 minute at 95°C, annealing for 1 minute at 58°C to 62°C (annealing temperatures optimized for each primer pair), and extension for 1.5 minutes at 72°C for 30 cycles followed by a terminal extension for 10 minutes at 72°C. A 2-μL sample of the final PCR product was diluted with 20 μL of stop solution (10 mmol/L EDTA, 0.1% sodium dodecyl sulfate [SDS], 95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol). After denaturation for 5 minutes at 95°C and chilling on ice for 5 minutes, 2 to 3 μL of each sample was electrophoresed through 6% polyacrylamide gels with 10% glycerol at 8 W to 10 W electricity for 16 hours to 20 hours at room temperature. Dried gels were autoradiographed at −80°C for 6 to 14 hours using intensifying screens.
Direct sequencing of PCR products. Tumor samples that showed an abnormal SSCP pattern were amplified for sequencing. Approximately 250 ng of DNA from these tumors was amplified as described earlier, but without isotope. The PCR product was purified through a Wizard PCR-prep spin column (Promega, Madison, WI) and sequenced by the dideoxy chain-termination method using an AmpliCycle sequencing kit (Perkin Elmer, Branchburg, NJ). Briefly, 10 pmol of one of the primers used for PCR amplification was end-labeled with γ[32P]dATP using T4 polynucleotide kinase. Approximately 10 ng of PCR-amplified DNA purified as described was mixed with 1 pmol of end-labeled primer, 4 μL of 10× cycle sequencing mixture, and 24 μL of sterile distilled water. Six microliters of this reaction mixture was mixed with 2 μL of each of the four dideoxy nucleotides, covered with 10 μL of mineral oil. The sequencing reaction was performed under the following conditions: initial denaturation for 5 minutes at 95°C was followed by 19 cycles of denaturation for 1 minute at 95°C, annealing for 1 minute at 62°C, and extension for 1 minute at 72°C. At the end of the reaction, 5 μL of stop buffer was added. Both strands were sequenced for each DNA fragment analyzed and verified by sequencing the product of two independent amplifications.
In four patients, direct sequencing analysis gave an ambiguous sequence ladder (with exon 5 primers in 3 patients and with exon 8 primers in 1 patient), probably due to the presence of either a deletion or a duplication in one of the alleles. To characterize these changes further, the PCR product from the amplification of genomic DNA from each of the five tumors was cloned into a TA-cloning vector (Invitrogen, San Diego, CA). Several recombinant clones were first screened by SSCP analysis to distinguish the normal allele from the abnormal allele. At least three independent clones for the abnormal allele and four clones for the normal allele were sequenced to identify the type of the abnormality. The abnormality was further confirmed by sequencing the second strand in each case.
Immunohistochemical studies for p53 expression. All tissues used for this study were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections at 4 to 5 μm were cut from each block and mounted on glass slides coated with poly-L-lysine. After dewaxing overnight in xylene, sections were rehydrated in graded alcohols, quenched in 3% hydrogen peroxide for 10 minutes, and washed in running distilled water. The wet slides were immersed in a plastic couplin jar filled with tissue-unmasking fluid (Signet, Dedham, MA) and heated in a microwave three times for 5 minutes each, with an interval of 1 minute between, at 600-W power. After cooling for 15 minutes in the same buffer, slides were washed in running distilled water and rinsed in phosphate-buffered saline (PBS).42 Each tissue section was covered with 300 to 400 μL of normal goat serum (1:10 dilution), incubated for 30 minutes at room temperature, and washed in PBS. Approximately 300 μL of primary p53 antibody (DAKO N1581; DAKO Corp, Carpinteria, CA) was applied and incubated overnight at 4°C. Slides were washed in PBS at room temperature and were treated with 400 μL of a secondary envision system (DAKO K1393) for 30 minutes at room temperature and washed in PBS. Finally, slides were covered with 400 μL of diaminobenzedene tetrachloride (DAB) for 4 minutes and washed in running water, counterstained with hematoxylin, dehydrated, and mounted.
For each tissue, the results were confirmed by repeating on two different slides, and all positive cases were confirmed by repeating the study a second time on an additional two slides. As a negative control, sections that had not been incubated with primary antibody were used. As a positive control, sections from a case of ovarian carcinoma showing p53 overexpression were used. The results were quantified as negative (−; no staining observed in any cell), positive (+) if 10% to 25% cells were positive, ++ if 25% to 50% cells were positive, and +++ if greater than 50% cells were positive.
Statistical significance of the data was tested by contingency chi-square analysis or by Fisher's exact method. Data on the duration of survival (from the date of diagnosis to the date of death or last follow-up evaluation) were examined by the product-limit method and compared using the log-rank test.43
RESULTS
In addition to histologic diagnosis, each of the 237 tumors included in this study were characterized by immunophenotypic analysis, immunogenotypic analysis, and cytogenetic analysis. Monoclonality for light-chain restriction and clonal rearrangements in the DNA of Ig heavy chain and either kappa or lambda light chain confirmed the B-cell origin of these tumors.
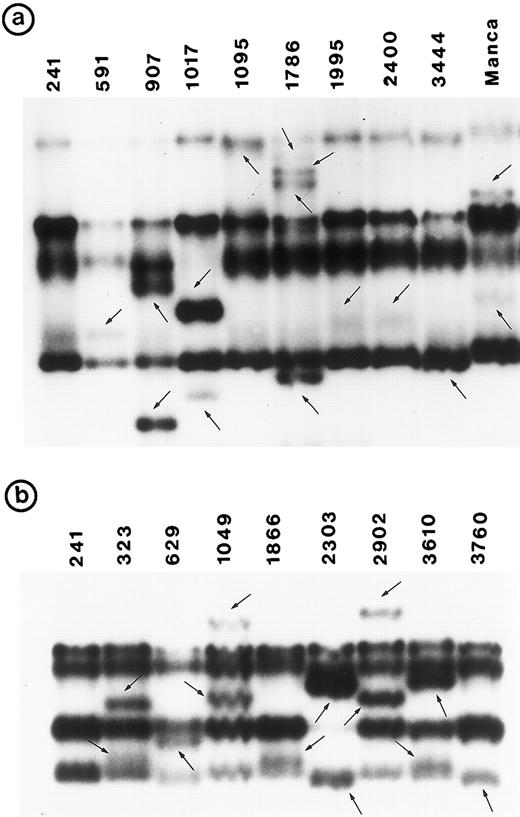
Southern blot analysis using the cDNA probe did not detect any gross alterations in P53 in any of these tumors. SSCP analysis showed abnormal conformation of the PCR products from exon 5 through exon 8 in 34 tumors (Fig 1). No change was identified in exon 9 (data not shown). Nucleotide sequence analysis of the PCR product from these 34 tumors showed that the abnormal SSCP conformation(s) was due to single nucleotide substitution in 24 tumors, deletion in three tumors, and duplication in one tumor (Table 1). The remaining six abnormal SSCP conformations were due to polymorphism in codon 213 (CGAarg → CGGarg ) in exon 6. Detailed analysis of histologic types is presented.
PCR-SSCP pattern of mutant P53 allele in B-NHL. Tumor genomic DNA was amplified with exon-specific primers and electrophoresed through nondenaturing polyacrylamide gels. (a) Exon 5, (b) exon 6, (c) exon 7, and (d) exon 8. Nongermline bands identified by arrows.
PCR-SSCP pattern of mutant P53 allele in B-NHL. Tumor genomic DNA was amplified with exon-specific primers and electrophoresed through nondenaturing polyacrylamide gels. (a) Exon 5, (b) exon 6, (c) exon 7, and (d) exon 8. Nongermline bands identified by arrows.
Correlation Between Cytogenetic Status of Chromosome 17, Mutation in P53, Protein Expression, and Duration of Survival in Different Histologic Types of B-NHL
| Tumor No. . | Histologic Type . | Chromosome 17p Status* . | Codon . | Codon Change . | Amino Acid Change . | Protein Expression† . | Survival (mo) . |
|---|---|---|---|---|---|---|---|
| 1095 | SLL | Normal | 147-153 | 21-bp duplication | Frame shift | + | NA |
| 323 | FSCC | Normal | 215 | AGT → ATT | Ser → Isol | ||
| 261 | AGG → AGT | Arg → Ser | + | T/60 | |||
| 840 | FSCC | Loss | 234 | TAC → AAC | Tyr → Asp | + | ?D/88 |
| 907 | FMx | Loss | 150-154 | 13-bp deletion | Frame shift | − | NA |
| 1786 | FMx | Deletion | 131 | 3-bp deletion | Frame shift | − | D/50+ |
| 459 | FLC | Abnormal | 241 | TCC → CCC | Ser → Pro | + | NA |
| 2303 | DSCC | Not done | 194 | CTT → CGT | Leu → Arg | + | D/5 |
| 1272 | DMx | Normal | 280 | AGA → ACA | Arg → Thr | +++ | D/NA |
| 1505 | DMx | Normal | 273 | CGT → TGT | Arg → Cys | + | D/10 |
| 541 | DLC | Normal | 248 | CGG → CAG | Arg → Gln | + | NA |
| 591 | DLC | Deleted | 145 | CTG → CCG | Leu → Pro | +++ | NA |
| 629 | DLC | Failure | 205 | TAT → TGT | Tyr → Cys | ||
| 239 | AAC → GAC | Asn → Asp | +++ | D/24 | |||
| 834 | DLC | Loss | 282 | CGG → TGG | Arg → Trp | + | NA |
| 919 | DLC | Failure | 240 | AGT → AGG | Ser → Arg | +++ | D/4 |
| 1017 | DLC | Failure | 175 | CGC → CAC | Arg → His | ++ | D/6 |
| 1049 | DLC | Not done | 220 | TAT → TGT | Tyr → Cys | − | D/14 |
| 1765 | DLC | Loss | 272 | GTG → GAG | Val → Glu | − | D/6 |
| 1866 | DLC | Normal | 213 | CGA → TGA | Arg → Stop | − | D/NA |
| 1995‡ | DLC | Deletion | 132 | AAG → AAC | Lys → Asp | +++ | D/21 |
| 2117 | DLC | Abnormal | 237 | ATG → ATA | Met → Isol | + | T/89+ |
| 2400‡ | DLC | Deletion | 132 | AAG → AAC | Lys → Asp | +++ | |
| 2432 | DLC | Normal | 288-289 | 2-bp deletion | Frame shift | ++ | D/10 |
| 2590 | DLC | Normal | 248 | CGG → CAG | Arg → Gln | +++ | D/24 |
| 2902 | DLC | Failure | 220 | TAT → TCT | Tyr → Ser | ++ | T/229+ |
| 3444 | DLC | Not done | 132 | AAG → GAG | Lys → Glu | ++ | D/28 |
| 3610 | DLC | Loss | 196 | CGA → TGA | Arg → stop | − | D/1 |
| Tumor No. . | Histologic Type . | Chromosome 17p Status* . | Codon . | Codon Change . | Amino Acid Change . | Protein Expression† . | Survival (mo) . |
|---|---|---|---|---|---|---|---|
| 1095 | SLL | Normal | 147-153 | 21-bp duplication | Frame shift | + | NA |
| 323 | FSCC | Normal | 215 | AGT → ATT | Ser → Isol | ||
| 261 | AGG → AGT | Arg → Ser | + | T/60 | |||
| 840 | FSCC | Loss | 234 | TAC → AAC | Tyr → Asp | + | ?D/88 |
| 907 | FMx | Loss | 150-154 | 13-bp deletion | Frame shift | − | NA |
| 1786 | FMx | Deletion | 131 | 3-bp deletion | Frame shift | − | D/50+ |
| 459 | FLC | Abnormal | 241 | TCC → CCC | Ser → Pro | + | NA |
| 2303 | DSCC | Not done | 194 | CTT → CGT | Leu → Arg | + | D/5 |
| 1272 | DMx | Normal | 280 | AGA → ACA | Arg → Thr | +++ | D/NA |
| 1505 | DMx | Normal | 273 | CGT → TGT | Arg → Cys | + | D/10 |
| 541 | DLC | Normal | 248 | CGG → CAG | Arg → Gln | + | NA |
| 591 | DLC | Deleted | 145 | CTG → CCG | Leu → Pro | +++ | NA |
| 629 | DLC | Failure | 205 | TAT → TGT | Tyr → Cys | ||
| 239 | AAC → GAC | Asn → Asp | +++ | D/24 | |||
| 834 | DLC | Loss | 282 | CGG → TGG | Arg → Trp | + | NA |
| 919 | DLC | Failure | 240 | AGT → AGG | Ser → Arg | +++ | D/4 |
| 1017 | DLC | Failure | 175 | CGC → CAC | Arg → His | ++ | D/6 |
| 1049 | DLC | Not done | 220 | TAT → TGT | Tyr → Cys | − | D/14 |
| 1765 | DLC | Loss | 272 | GTG → GAG | Val → Glu | − | D/6 |
| 1866 | DLC | Normal | 213 | CGA → TGA | Arg → Stop | − | D/NA |
| 1995‡ | DLC | Deletion | 132 | AAG → AAC | Lys → Asp | +++ | D/21 |
| 2117 | DLC | Abnormal | 237 | ATG → ATA | Met → Isol | + | T/89+ |
| 2400‡ | DLC | Deletion | 132 | AAG → AAC | Lys → Asp | +++ | |
| 2432 | DLC | Normal | 288-289 | 2-bp deletion | Frame shift | ++ | D/10 |
| 2590 | DLC | Normal | 248 | CGG → CAG | Arg → Gln | +++ | D/24 |
| 2902 | DLC | Failure | 220 | TAT → TCT | Tyr → Ser | ++ | T/229+ |
| 3444 | DLC | Not done | 132 | AAG → GAG | Lys → Glu | ++ | D/28 |
| 3610 | DLC | Loss | 196 | CGA → TGA | Arg → stop | − | D/1 |
Abbreviations: D, specimen studied at diagnosis; T, specimen studied after transformation; NA, data not available.
Loss, monosomy; deletion, deletion of short arm.
+, 10% to 25% cells positive; ++, 26% to 50% cells positive; +++, >51% cells positive.
Specimens obtained from the same patient at different times.
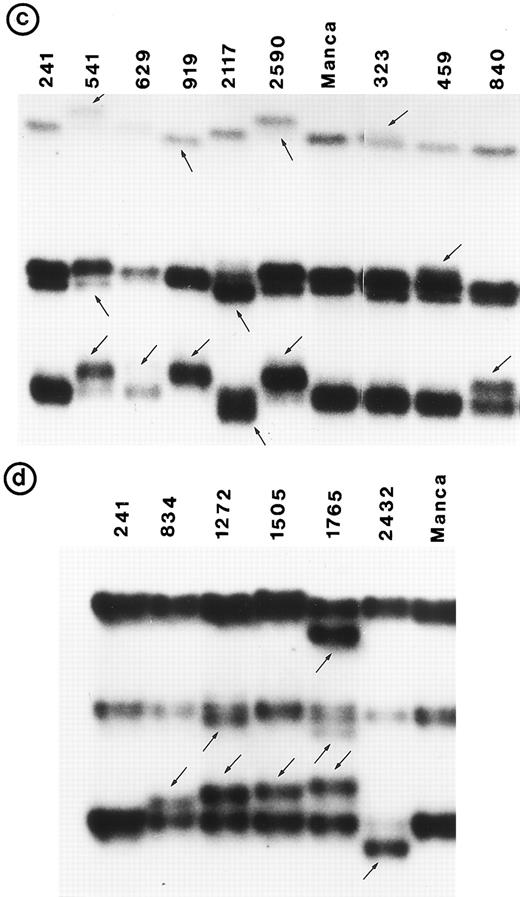
Small lymphocytic lymphoma (SLL; 45 tumors). Cytogenetic studies were performed on 41 tumors, of which only 22 had clonal karyotypic abnormalities; 7 tumors had only normal karyotype and no metaphases were obtained in 12 tumors. Chromosome 17 was abnormal in 3 tumors due to monosomy in 2 and to a derived 17p in 1; however, no mutation was identified in these 3 tumors. One other tumor (no. 1095) in which chromosome 17p was apparently normal had a tandem duplication of 21 bp in exon 5 (Fig 2b).
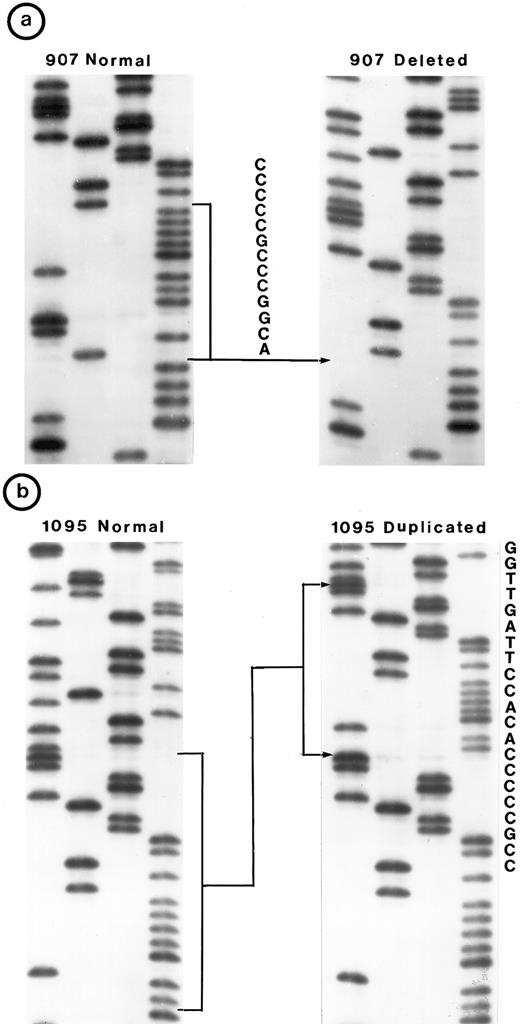
Nucleotide sequence of representative mutant P53 alleles. DNA from tumors scored positive on PCR-SSCP analysis were further characterized by nucleotide sequencing of both DNA strands. (a and b) Structural changes in exon 5. PCR product from these tumors was cloned into a TA-vector from which normal and mutant alleles were isolated and sequenced. Tumor no. 907 had a deletion and no. 1095 had a duplication.
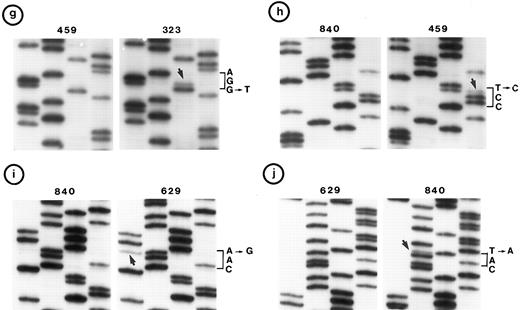
(c, d, e, and f) Representative point mutations in exon 6. (g, h, i, and j) Representative point mutations in exon 7. PCR-amplified DNA from these tumors was sequenced directly. Note that both homologs of chromosome 17 were cytogenetically normal in nos. 323, 1095, and 1866; 1 copy of 17p was lost in nos. 459, 840, and 905; status of chromosome 17 was not known in nos. 629 and 2303.
Nucleotide sequence of representative mutant P53 alleles. DNA from tumors scored positive on PCR-SSCP analysis were further characterized by nucleotide sequencing of both DNA strands. (a and b) Structural changes in exon 5. PCR product from these tumors was cloned into a TA-vector from which normal and mutant alleles were isolated and sequenced. Tumor no. 907 had a deletion and no. 1095 had a duplication.
(c, d, e, and f) Representative point mutations in exon 6. (g, h, i, and j) Representative point mutations in exon 7. PCR-amplified DNA from these tumors was sequenced directly. Note that both homologs of chromosome 17 were cytogenetically normal in nos. 323, 1095, and 1866; 1 copy of 17p was lost in nos. 459, 840, and 905; status of chromosome 17 was not known in nos. 629 and 2303.
Follicular small cleaved-cell lymphoma (FSCC; 38 tumors). Clonal karyotypic abnormalities were identified in 31 of 36 tumors studied cytogenetically. Two tumors had monosomy for chromosome 17; one of these (no. 840) had a mutation in exon 7. On the other hand, one other tumor (no. 323) with apparently normal homologs of chromosome 17 had two mutations: one in exon 6 and the other in exon 7 (Fig 2c and g).
Follicular mixed small cleaved-cell and large-cell lymphoma (FMx; 35 tumors). Clonal cytogenetic abnormalities were identified in all 31 tumors studied. Monosomy for chromosome 17 was present in three tumors; one of these (no. 907) had a 13-bp deletion in exon 5 (Fig 2a). One tumor had del(17p) and a deletion of 3 bp in exon 5.
Follicular large-cell lymphoma (FLC; 4 tumors). Clonal cytogenetic abnormalities were identified in three tumors. One tumor (no. 459) had an abnormal 17p due to a derived chromosome. This tumor had a mutation in exon 7 (Fig 2h).
Diffuse small cleaved-cell lymphoma (DSCC; 14 tumors). Cytogenetic studies were performed in 11 tumors; 7 tumors had clonal karyotypic abnormalities. Chromosome 17 abnormalities were not identified in these tumors. One tumor (no. 2303), which was not analyzed cytogenetically, had a mutation in exon 6 (Fig 2e).
Diffuse mixed small- and large-cell lymphoma (DMx; 17 tumors). Clonal karyotypic abnormalities were identified in 15 of 16 tumors cytogenetically studied. One tumor had an i(17q), but no mutation was identified in P53. On the other hand, two tumors (no. 1272 and 1505) with apparently normal homologs for chromosome 17 had a mutation in exon 8.
DLC (84 tumors). Cytogenetic studies were performed in 75 tumors obtained from 74 patients; of these, clonal karyotypic abnormalities were identified in 59 tumors obtained from 58 patients. Chromosome 17 was abnormal in 15 tumors (20%) due to monosomy in 5 tumors, to a der(17p) chromosome in 3, to a del(17p) in 6, and to an i(17q) in 1.
A mutation in P53 was identified in 6 patients (7 tumors) with abnormalities of chromosome 17p and in 4 other tumors in which chromosome 17 was cytogenetically normal. In addition to these, a mutation in P53 was identified in 6 other tumors for which cytogenetic data were not obtained; one of these (no. 629) had two mutations (Fig 2f and i). Among these 17 mutations, 16 were single nucleotide substitutions and one was a 2-bp deletion (Table 1). In two tumors, point mutations generated a premature stop signal (Fig 2d). Tumors no. 1995 and no. 2400 were obtained from the same patient at different times; both had a similar karyotype and showed an identical mutation in exon 5.
Mantle-cell lymphoma (10 tumors). Among 13 patients with a t(11; 14)(q13; q32), 10 tumors were identified as mantle-cell lymphomas. A mutation in P53 was identified in three tumors and three of six tumors examined were positive for p53 by immunohistochemical staining.
Types of mutations. We detected 27 different somatic changes in exons 5 through 8 of P53 in 26 tumors from 25 patients with various histologic types of B-NHL; however, they were more frequent in lymphomas of DLC and mantle-cell histology than of the other types (P < .01). No mutation was identified in exon 9. In 20 tumors (from 19 patients), 21 different missense mutations led to a change in amino acid residue (Table 1). Codons for eight different amino acids, namely, Arg, Asn, Leu, Lys, Met, Ser, Tyr, and Val, were affected by these mutations; the codon for arginine was the most frequently affected (9 of 23). Mutations of nucleotide transition type occurred more frequently than transversion type (15:8). Analysis of the spectrum of single nucleotide substitutions in these tumors showed a high frequency of transition mutations in CpG dinucleotide sequences (7 of 23). In two tumors, a G → T transversion was present. A mutation in two different exons was identified in two tumors (no. 323 and 629). Whether they occurred in each allele of the gene, ie, compound heterozygosity, or in two exons of the same allele cannot be determined. Compound heterozygotes have been reported infrequently.44 45 In two tumors, the point mutation resulted in a premature stop signal; interestingly, both of these mutations were C → T transitions in the codon CGA for arginine. Four tumors had structural changes, namely, intraexonic deletion (3 tumors) or intraexonic duplication (1 tumor) (Table 1).
Cytogenetic complexity and mutation. Cytogenetic studies were performed on 216 of 237 tumors included in this study. Clonal karyotypic abnormalities were identified in 168 tumors (78%); 99 of them (59%) had complex karyotypes containing three or more structural abnormalities and 69 (41%) had simple abnormal karyotypes containing less than three structural changes.
Karyotypes were obtained from 19 of 26 tumors in which a mutation(s) was identified; all had complex cytogenetic changes (Table 2); in 11 of these tumors, 17p was abnormal (structural abnormality in 6, monosomy for 17 in 5). Cytogenetic studies were not performed for three tumors and no karyotypes were obtained from the cultures of four tumors.
Histologic, Immunologic, and Genetic Features of B-NHLs With Mutated P53
| Tumor No. . | Age/Sex . | Cell-Surface Markers . | Genotype . | Karyotype . | Mutated Exon . |
|---|---|---|---|---|---|
| SLL | |||||
| 1095 | 58/F | IgHM, IgLK | IgHr, IgKr | 46,XX[1]/46,XX,del(6)(q13q21),del(9)(q22q24),t(11; 14)(q13; q32)[14] | 5 |
| FSCC | |||||
| 323 | 39/M | IgHM, IgLK | IgHr, IgKr | 46,XY[2]/48,XY,+X,−5,add(9)(p13),der(10)t(7; 10)(p13; p13),add(12)(q13),t(4; 18)(q32; q21),+der(18)t(14; 18),+mar[15] | 6,7 |
| 840 | 65/M | IgHM, IgLL | IgHr, IgLr | 48,XY,t(3; 14)(q27; q32),del(8)(q24),t(12; 12)(q24; q13),+20,+mars[5]/89,XXYY,(4n+/−),−2,−3,−5,t(3; 14),−6,del(8),−9,t(12; 12),−14,−14,−17,−19,+20, +mars[2] | 7 |
| FMx | |||||
| 907 | 79/F | IgHM, IgLL | IgHr, IgKg, IgLr | 46,XX[3]/46,XX,t(3; 5)(q27; q13),t(3; 14)(q27; q32),del(5)(q11q33)[1]/80-82,XXX,del(X)(q22q26),+del(X),(4n+/−),der(1)t(1; 1)(p34; q23),t(3; 14)(q27; q32), der(3)t(3; 5)(p25; p13),−4,del(6)(q13q25),der(6)t(6; 11)(q25; q13),del(7)(p11),−8,der(9)t(9; 13)(p11; q12),add(9)(q34),−10,del(11)(q21q23),−13,−15, −15,−16,−17,add(19)(p13),der(19)t(?17; 19)(p13; q13),−20,−22[11] | 5 |
| 1786 | 52/M | IgHM, IgLK | IgHr, IgKr | 46,XY[8]/46,XY,+X,−2,r(3)(q25q27),del(4)(p14),add(4)(q31),−6,del(8)(q13q22),add(9)(p13),add(9)(q34),+11,del(17)(p11),−18,+mar[9]/46,XY,+X,−2, t(3; 9)(p13; q34),−4,−6,del(8),+11,del(17),−18,+mars[9] | 5 |
| FLC | |||||
| 459 | 58/F | IgHM, IgLK | IgHr, IgKr | 46,XX[4]/46,XX,dup(7)(pter→q34∷q34→q11∷q32→ter),−9,→ter),add(9)(p12),add(17)(p11),+18, +21, random loss[3] | 7 |
| DSCC | |||||
| 2303 | 76/F | IgHM, IgLK | IgHr, IgKr | Not done | 6 |
| DMx | |||||
| 1272 | 81/F | IgH−, IgLK | IgHr, IgKg | 53-56,XX,+X,+X,+4,−6,+7,t(8; 14)(q24; q32),+11,+12,+der(14)t(8; 14),der(16)t(1; 16)(q25; p13),+17,+21,+21,+mars,random loss[12] | 8 |
| 1505 | 62/M | IgHM, IgLL | IgHr, IgKr | 46,XY,ins(1)(pter→q21∷?∷q21→ter),t(3; 14)(q27; q11)[20] | 8 |
| DLC | |||||
| 541 | 87/F | IgM, IgLK | IgHr, IgKr | 42-48,XX,+dup(X)(qter→?p22∷q13→ter),t(1; 6)(p13; q13),+del(5)(q12),t(7; 12)(q34; q13),der(8)t(8; 9)(p11; p13),−10,−13,add(19)(q13),+add(19),add(20) (q13),+2xadd(20),−22,+mars[20] | 7 |
| 591 | 80/F | ND | IgHr, IgKr | 86-88,XXXX,(4n+/−),del(3)(q27),2xadd(4)(q31),2xdel(6)(q15q25),2xadd(6)(q23),−10,−14,−16,−17,del(17)(p11),−19,−19,+mars[10] | 5 |
| 629 | 40/F | ND | IgHr, IgKr | Failure | 6,7 |
| 834 | 75/F | IgH−, IgLL | IgHr, IgKr | 46,XX[2]/82-84,XXX,(4n+/−),−1,del(1p34),+add(3)(q37),+?t(3; 3)(q27; p21),−4,−4,−5,−6,−6,−6,+9,−14,−14,+15,−17,−17,−21,−21,+mars[12] | 8 |
| 919 | 60/M | IgHM, IgLK | IgHr, IgKr | Failure | 7 |
| 1017 | 57/M | IgHND, IgLL | IgHr, IgKr | Failure | 5 |
| 1049 | 71/M | ND | IgHr, IgKr | Not done | 6 |
| 1765 | 62/M | IgH−, IgLL | IgHr, IgKr | 46,XY[7]/41,X,−Y,del(1)(p13p34),−10,t(11; 14)(q13; q32),dup(11)(pter→q23∷q13→q23∷?),−12,−13,−17[13] | 8 |
| 1866 | 57/M | IgHM, IgLK | IgHr, IgKr | 46,XY[2]/47,XY,+X[1]/74-95,XXYY(3n+/−),del(1)(p13p36),del(1)(q21),+3,del(7)(q22q34),+del(7),+8,+11,del(13)(q13q22),+14,+16,+16,+17,−18,−19, +20,+21,+22,+mars[9] | 6 |
| 1995 | 83/F | IgHND, IgLL | IgHr, IgKr | 87-92,XXXX,(4n+/−),+X,+inv(X)(p11p22),−2,t(3; 22)(q37; q11),add(6)(q25),−8,−9,dup(12)(pter→q15∷q13→ter),−16,del(17)(p11),−20,+mars[18] | 5 |
| 2117 | 64/F | ND | IgHr, IgKr | 46,XX[1]/51,XX,+add(X)(p22),inv(3)(q13q27),+8,+8,+11,+11,t(14; 18)(q32; q21),add(17)(p11)[9] | 7 |
| 2400 | 84/F | ND | IgHr, IgKr | 46,XX[1]/87-92,XXXX,(4n+/−),+X,+inv(X)(p11p12),−2,−3,t(3; 22)(q27; q11),add(6)(q25),−6,−8,−8,−9,del(9)(p11),−10,−11,der(12)(pter→q15∷13→ter), t(13; 15)(q11; p11),−16,−17,del(17)(p11),−20,−20,+mars[18] | 5 |
| 2432 | 51/F | IgH−, IgLL | IgHr, IgKr | 45,X,add(X)(q21),add(1)(q23),add(2)(p11),del(3)(q13),t(3; 14)(q27; q32),i(8q),add(12)(q24),−15[6]/46,XX,t(2; 10)(q37; q11)[1] | 8 |
| 2590 | 66/F | IgHM, IgLK | IgHr, IgKr | 46,XX[9]/41-43,XX,−1,add(5)(q31),t(11; 14)(q13; q32),−13,+2xadd(19)(q13),+mar1,+mar2,random loss[11] | 7 |
| 2902 | 68/M | IgHND, IgLK | IgHr, IgKr | Failure | 6 |
| 3444 | 84/F | IgHND, IgLL | IgHr, IgKr | Not done | 5 |
| 3610 | 72/F | Ig− | IgHd, IgKd, IgLr | 46,XX[9]/76-93,XX,(4n+/−),−X,−X,−5,add(7)(p15),−9,der(9)t(9; 17)(p12; q11),2xadd(11)(q23),+12,−14,+16,−17,+18,+18,+18,+18,−22,+mars[11] | 6 |
| Abbreviations: IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; M, mu heavy chain; K, kappa light chain; L, lambda light chain; g, germline; r rearranged; d, deleted; ND, study not done due to insufficient material. |
| Tumor No. . | Age/Sex . | Cell-Surface Markers . | Genotype . | Karyotype . | Mutated Exon . |
|---|---|---|---|---|---|
| SLL | |||||
| 1095 | 58/F | IgHM, IgLK | IgHr, IgKr | 46,XX[1]/46,XX,del(6)(q13q21),del(9)(q22q24),t(11; 14)(q13; q32)[14] | 5 |
| FSCC | |||||
| 323 | 39/M | IgHM, IgLK | IgHr, IgKr | 46,XY[2]/48,XY,+X,−5,add(9)(p13),der(10)t(7; 10)(p13; p13),add(12)(q13),t(4; 18)(q32; q21),+der(18)t(14; 18),+mar[15] | 6,7 |
| 840 | 65/M | IgHM, IgLL | IgHr, IgLr | 48,XY,t(3; 14)(q27; q32),del(8)(q24),t(12; 12)(q24; q13),+20,+mars[5]/89,XXYY,(4n+/−),−2,−3,−5,t(3; 14),−6,del(8),−9,t(12; 12),−14,−14,−17,−19,+20, +mars[2] | 7 |
| FMx | |||||
| 907 | 79/F | IgHM, IgLL | IgHr, IgKg, IgLr | 46,XX[3]/46,XX,t(3; 5)(q27; q13),t(3; 14)(q27; q32),del(5)(q11q33)[1]/80-82,XXX,del(X)(q22q26),+del(X),(4n+/−),der(1)t(1; 1)(p34; q23),t(3; 14)(q27; q32), der(3)t(3; 5)(p25; p13),−4,del(6)(q13q25),der(6)t(6; 11)(q25; q13),del(7)(p11),−8,der(9)t(9; 13)(p11; q12),add(9)(q34),−10,del(11)(q21q23),−13,−15, −15,−16,−17,add(19)(p13),der(19)t(?17; 19)(p13; q13),−20,−22[11] | 5 |
| 1786 | 52/M | IgHM, IgLK | IgHr, IgKr | 46,XY[8]/46,XY,+X,−2,r(3)(q25q27),del(4)(p14),add(4)(q31),−6,del(8)(q13q22),add(9)(p13),add(9)(q34),+11,del(17)(p11),−18,+mar[9]/46,XY,+X,−2, t(3; 9)(p13; q34),−4,−6,del(8),+11,del(17),−18,+mars[9] | 5 |
| FLC | |||||
| 459 | 58/F | IgHM, IgLK | IgHr, IgKr | 46,XX[4]/46,XX,dup(7)(pter→q34∷q34→q11∷q32→ter),−9,→ter),add(9)(p12),add(17)(p11),+18, +21, random loss[3] | 7 |
| DSCC | |||||
| 2303 | 76/F | IgHM, IgLK | IgHr, IgKr | Not done | 6 |
| DMx | |||||
| 1272 | 81/F | IgH−, IgLK | IgHr, IgKg | 53-56,XX,+X,+X,+4,−6,+7,t(8; 14)(q24; q32),+11,+12,+der(14)t(8; 14),der(16)t(1; 16)(q25; p13),+17,+21,+21,+mars,random loss[12] | 8 |
| 1505 | 62/M | IgHM, IgLL | IgHr, IgKr | 46,XY,ins(1)(pter→q21∷?∷q21→ter),t(3; 14)(q27; q11)[20] | 8 |
| DLC | |||||
| 541 | 87/F | IgM, IgLK | IgHr, IgKr | 42-48,XX,+dup(X)(qter→?p22∷q13→ter),t(1; 6)(p13; q13),+del(5)(q12),t(7; 12)(q34; q13),der(8)t(8; 9)(p11; p13),−10,−13,add(19)(q13),+add(19),add(20) (q13),+2xadd(20),−22,+mars[20] | 7 |
| 591 | 80/F | ND | IgHr, IgKr | 86-88,XXXX,(4n+/−),del(3)(q27),2xadd(4)(q31),2xdel(6)(q15q25),2xadd(6)(q23),−10,−14,−16,−17,del(17)(p11),−19,−19,+mars[10] | 5 |
| 629 | 40/F | ND | IgHr, IgKr | Failure | 6,7 |
| 834 | 75/F | IgH−, IgLL | IgHr, IgKr | 46,XX[2]/82-84,XXX,(4n+/−),−1,del(1p34),+add(3)(q37),+?t(3; 3)(q27; p21),−4,−4,−5,−6,−6,−6,+9,−14,−14,+15,−17,−17,−21,−21,+mars[12] | 8 |
| 919 | 60/M | IgHM, IgLK | IgHr, IgKr | Failure | 7 |
| 1017 | 57/M | IgHND, IgLL | IgHr, IgKr | Failure | 5 |
| 1049 | 71/M | ND | IgHr, IgKr | Not done | 6 |
| 1765 | 62/M | IgH−, IgLL | IgHr, IgKr | 46,XY[7]/41,X,−Y,del(1)(p13p34),−10,t(11; 14)(q13; q32),dup(11)(pter→q23∷q13→q23∷?),−12,−13,−17[13] | 8 |
| 1866 | 57/M | IgHM, IgLK | IgHr, IgKr | 46,XY[2]/47,XY,+X[1]/74-95,XXYY(3n+/−),del(1)(p13p36),del(1)(q21),+3,del(7)(q22q34),+del(7),+8,+11,del(13)(q13q22),+14,+16,+16,+17,−18,−19, +20,+21,+22,+mars[9] | 6 |
| 1995 | 83/F | IgHND, IgLL | IgHr, IgKr | 87-92,XXXX,(4n+/−),+X,+inv(X)(p11p22),−2,t(3; 22)(q37; q11),add(6)(q25),−8,−9,dup(12)(pter→q15∷q13→ter),−16,del(17)(p11),−20,+mars[18] | 5 |
| 2117 | 64/F | ND | IgHr, IgKr | 46,XX[1]/51,XX,+add(X)(p22),inv(3)(q13q27),+8,+8,+11,+11,t(14; 18)(q32; q21),add(17)(p11)[9] | 7 |
| 2400 | 84/F | ND | IgHr, IgKr | 46,XX[1]/87-92,XXXX,(4n+/−),+X,+inv(X)(p11p12),−2,−3,t(3; 22)(q27; q11),add(6)(q25),−6,−8,−8,−9,del(9)(p11),−10,−11,der(12)(pter→q15∷13→ter), t(13; 15)(q11; p11),−16,−17,del(17)(p11),−20,−20,+mars[18] | 5 |
| 2432 | 51/F | IgH−, IgLL | IgHr, IgKr | 45,X,add(X)(q21),add(1)(q23),add(2)(p11),del(3)(q13),t(3; 14)(q27; q32),i(8q),add(12)(q24),−15[6]/46,XX,t(2; 10)(q37; q11)[1] | 8 |
| 2590 | 66/F | IgHM, IgLK | IgHr, IgKr | 46,XX[9]/41-43,XX,−1,add(5)(q31),t(11; 14)(q13; q32),−13,+2xadd(19)(q13),+mar1,+mar2,random loss[11] | 7 |
| 2902 | 68/M | IgHND, IgLK | IgHr, IgKr | Failure | 6 |
| 3444 | 84/F | IgHND, IgLL | IgHr, IgKr | Not done | 5 |
| 3610 | 72/F | Ig− | IgHd, IgKd, IgLr | 46,XX[9]/76-93,XX,(4n+/−),−X,−X,−5,add(7)(p15),−9,der(9)t(9; 17)(p12; q11),2xadd(11)(q23),+12,−14,+16,−17,+18,+18,+18,+18,−22,+mars[11] | 6 |
| Abbreviations: IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; M, mu heavy chain; K, kappa light chain; L, lambda light chain; g, germline; r rearranged; d, deleted; ND, study not done due to insufficient material. |
A mutation in P53 was identified in 19 of 99 tumors with complex cytogenetic abnormalities; whereas no mutation was identified in 69 tumors with simple cytogenetic changes; the difference was significant (P < .001). Furthermore, a mutation in P53 was identified in a significantly higher proportion of tumors with an abnormality of chromosome 17 (11 of 25 tumors) than in tumors with an apparently normal chromosome 17 (8 of 143 tumors) (P < .0001).
p53 expression. Expression of p53 was studied by immunochemical staining on histologic sections from formalin-fixed tissue blocks of 173 patients (174 tumors) separated into three different groups, namely, P53 mutants, nonmutants with 17p abnormalities, and nonmutants with cytogenetically normal homologs of 17. There was some variation in the intensity of staining (weak to strong); however, staining was discrete and was localized to nuclei. In some tumors of all histologic groups, strongly positive isolated cells were observed; however, these tumors were scored as negative for the purpose of this study. Excluding the latter type, 81 tumors were scored positive for p53.
In the first group of 26 tumors with a mutation in P53, 20 tumors (19 patients) were positive for p53. This is in contrast to the frequency of 61 p53-positive tumors identified among 147 tumors without a mutation; the difference was significant (P < .001). Furthermore, p53 was detected in a significantly higher proportion of tumors with a missense mutation (17 of 19) than in tumors with a nonsense mutation (2 of 6; P < .05). Among six p53-negative tumors, two had a point mutation that generated a premature stop codon, two had a deletion, and the remaining two had missense mutations (Table 1).
In the second group, immunostaining for p53 was performed on 12 of 14 tumors with a clonal cytogenetic abnormality affecting 17p. In seven tumors, moderate to intense staining was noted and five tumors did not show staining (Table 3). This frequency was significantly higher when compared with tumors with apparently normal 17 in which 61 of 147 tumors were p53-positive (P < .05).
Variation in the Expression of p53 in Lymphoma Tumors Without a Mutation in Exons 5 Through 9
| % p53-Positive Cells . | Histologic Type . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | SLL . | FSCC . | FMx . | FLC . | DSCC . | DMx . | DLC . | Total . | . | . | . | . | . | . | . | . | ||||||||
| . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | . | . | . | . | . | . | . | . |
| None | 1 | 25 | 2 | 17 | ND | 11 | ND | ND | 0 | 9 | 1 | 9 | 1 | 11 | 5 | 82 | ||||||||
| 10% to25% | 1 | 6 | 0 | 6 | ND | 8 | ND | ND | 1 | 3 | 0 | 2 | 0 | 2 | 2 | 27 | ||||||||
| 26% to 50% | 0 | 2 | 0 | 2 | ND | 3 | ND | ND | 0 | 0 | 0 | 2 | 3 | 7 | 3 | 16 | ||||||||
| >50% | 0 | 1 | 0 | 1 | ND | 2 | ND | ND | 0 | 1 | 0 | 1 | 2 | 5 | 2 | 11 | ||||||||
| Total | 2 | 34 | 2 | 26 | ND | 24 | ND | ND | 1 | 13 | 0 | 14 | 6 | 25 | 12 | 136 | ||||||||
| % p53-Positive Cells . | Histologic Type . | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | SLL . | FSCC . | FMx . | FLC . | DSCC . | DMx . | DLC . | Total . | . | . | . | . | . | . | . | . | ||||||||
| . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | 1 . | 2 . | . | . | . | . | . | . | . | . |
| None | 1 | 25 | 2 | 17 | ND | 11 | ND | ND | 0 | 9 | 1 | 9 | 1 | 11 | 5 | 82 | ||||||||
| 10% to25% | 1 | 6 | 0 | 6 | ND | 8 | ND | ND | 1 | 3 | 0 | 2 | 0 | 2 | 2 | 27 | ||||||||
| 26% to 50% | 0 | 2 | 0 | 2 | ND | 3 | ND | ND | 0 | 0 | 0 | 2 | 3 | 7 | 3 | 16 | ||||||||
| >50% | 0 | 1 | 0 | 1 | ND | 2 | ND | ND | 0 | 1 | 0 | 1 | 2 | 5 | 2 | 11 | ||||||||
| Total | 2 | 34 | 2 | 26 | ND | 24 | ND | ND | 1 | 13 | 0 | 14 | 6 | 25 | 12 | 136 | ||||||||
Frequency of tumors showing variable expression of p53 in tumors (1) with abnormality affecting 17p, and (2) with apparently normal 17p.
Abbreviation: ND, immunostaining not performed.
In the third group, expression of p53 was studied in 136 tumors from different histologic groups; 54 tumors were positive for p53 (Table 3) and the majority of them had a mixture of p53-negative and p53-positive cells homogeneously distributed (Fig 3). In the FMx, DLC, and MC histologic groups, more than 50% of the tumors examined were positive for p53. In DLC tumors, there was no significant difference in the frequency of p53-positive and p53-negative patients with or without mutations. No significant association was observed between cytogenetic complexity and p53 expression; thus, 26 of 42 p53-positive tumors and 20 of 47 p53-negative tumors had complex cytogenetic abnormalities. However, among tumors with greater than 25% p53-positive cells, 10 of 15 tumors had complex cytogenetic abnormalities, whereas only four of 25 tumors with less than 25% p53-positive cells had complex cytogenetic abnormalities; the difference was significant (P < .01).
Representative examples of p53 expression in B-NHL. Sections from formlin-fixed tissues were incubated with DAKO N1581 p53 antibody. Tumor cells (nuclei) expressing p53 were stained brown. (A) SLL, (B) FSCC, (C) a portion of B as viewed under a higher magnification, (D) FMx, (E) DMx, (F) DCL.
Representative examples of p53 expression in B-NHL. Sections from formlin-fixed tissues were incubated with DAKO N1581 p53 antibody. Tumor cells (nuclei) expressing p53 were stained brown. (A) SLL, (B) FSCC, (C) a portion of B as viewed under a higher magnification, (D) FMx, (E) DMx, (F) DCL.
P53 mutation and survival. In general, among nonmutants, 22 of 49 patients died within 1 to 68 months (median, 13), whereas 14 of 17 patients with a mutation died within 1 to 88 months (median, 12; P < .05). In 15 patients, a mutation in P53 was identified at diagnosis; data on survival were available from 13 of these patients (Table 1). One patient (no. 1786) had no evidence of disease 50 months after diagnosis. The remaining 12 patients died within 1 to 28 months (median, 10). Patient no. 840 was diagnosed with FSCC lymphoma in 1985. A mutation in P53 was identified in a specimen obtained in September 1990; this also showed FSCC lymphoma. This patient died 88 months after diagnosis with no evidence of histologic progression. Patient no. 323 had FSCC histology at diagnosis (January 1986). A biopsy obtained in August 1989, in which a mutation in P53 was identified, also showed FSCC histology. Disease progressed subsequently to DLC histology and the patient died 60 months after the initial diagnosis. The other two patients (nos. 2117 and 2902) transformed from FSCC to DLC and a mutation was identified after histologic transformation. No material from the initial tumor was available for analysis in either of these patients. Both patients had no evidence of disease 89 months and 229 months after the initial diagnosis, respectively. Sufficient clinical data on initial diagnosis were not available in the remaining six patients with a mutation in P53.
P53 mutation, p53 expression, and survival. p53 was expressed in 19 of 25 patients with a mutation; two were alive and 11 died within 4 to 88 months (median, 14). Among six patients with a mutation, but not p53, one was alive at 53 months and three died within 1 to 14 months (median, 6; P > .3). In the p53-negative patients, a significant difference was found in the number of survivors and nonsurvivors between mutants and nonmutants. Thus, three of four mutants and one of 24 nonmutants died (P < .01). In DLC patients with a mutation, two of nine p53-positive and none of three p53-negative patients survived; however, the difference was not significant (P > .5).
p53 expression and survival. In another group of 50 patients without a mutation, the relationship between p53 expression and survival was examined. Among 26 p53-positive patients, 21 died within 1 to 68 months (median, 13) and five were alive at a median follow-up duration of 33 months (range, 9 to 66); on the other hand, among p53-negative patients, one patient died at 63 months and 23 were alive at a median follow-up duration of 46 months (range, 28 to 116). The difference between the two groups was significant (P < .001). In patients with FSCC tumors, four of five p53-positive patients died, as opposed to none among nine p53-negative patients (P < .005). Similarly, in patients with DLC tumors, 15 of 17 p53-positive patients died within 3 to 68 months (median, 17), and one of eight p53-negative patients died at 63 months and seven were alive at a median follow-up duration of 58 months (range, 29 to 89; P < .001).
Cytogenetic complexity and survival. The relationship between cytogenetic complexity and survival was also examined in patients with and without a mutation. Among patients with normal P53, 12 with complex cytogenetic abnormalities died within 5 to 68 months (median, 22) and 13 survived at a median follow-up duration of 38 months (range, 23 to 116). On the other hand, all nine patients with simple cytogenetic abnormalities survived at a median follow-up duration of 48 months (range, 29 to 67; P < .05). No significant difference was noted in the frequency of survivors and nonsurvivors among patients with complex and simple cytogenetic changes in DLC histologic group (data not shown).
DISCUSSION
The p53 transcription regulatory protein46 monitors the integrity47 of the genome by arresting cells at G1 or programming them to cell death when DNA replication is defective or when DNA is damaged.48-51 Therefore, inactivation of this gene would provide a selective advantage for clonal expansion of neoplastic cells. Furthermore, this would be particularly significant in determining the effect of therapeutic agents that cause DNA damage. Mutant P53 represents a loss of function by a recessive or dominant negative process.47 Previous studies have shown that majority of mutations occur as missense changes in the highly conserved sequences of the coding region (exons 5 through 8).2
Previous studies of P53 changes in lymphoma were limited to selected histologic types, whereas we examined the spectrum of somatic changes in 237 unselected B-NHLs to assess their association with different histologic types and clinical significance. We used a combination of cytogenetics, Southern blotting, SSCP analysis, direct sequencing, and p53 expression methods. Therefore, we believe that we have identified most of the somatic changes in P53. Nonetheless, this study cannot exclude mutations not covered by the primer sets used for exons 5 through 9 and/or those mutations that do not lead to overexpression of protein. We identified a mutation rate of 10.5% in P53 in B-NHL in general and a 19% mutation rate for DLC in particular. Mutations were identified in tumors at diagnosis and after histologic conversion. These findings are in contrast to previous reports that mutations in P53 were late-stage events and were present in advanced histologic groups or in tumors after histologic conversion or relapse.18,29,41,52 53
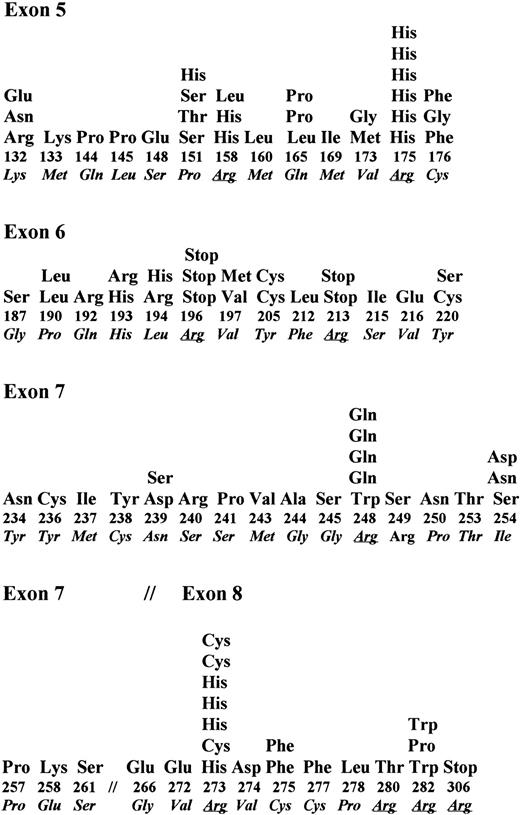
In B-NHL, somatic changes in exons 5 through 8 of P53 have been identified in 108 of 536 tumors analyzed in this study and others.17,18,29,30,32,40,52-57 Single nucleotide substitutions were identified in 96 tumors, deletion of 1 bp to 11 bp was identified in 11 tumors, and a duplication in exon 5 was identified in one tumor. Nucleotide substitutions of transition type occurred predominantly (58%) over transversion type (37.5%). Four tumors showed two point mutations in the same codon. A high proportion of the mutations occurred at the CpG dinucleotide sequences (27 of 96 mutations). Among the different exons examined, 13 of 61 codons in exon 5 were affected by 30 mutations (mutation frequency of 0.21/codon), 13 of 38 codons of exon 6 were affected by 23 mutations (mutation frequency of 0.34/codon), 13 of 37 codons of exon 7 were affected by 24 mutations (mutation frequency of 0.35/codon), and 10 of 45 codons of exon 8 were affected by 19 mutations (mutation frequency of 0.22/codon). Thus, exons 6 and 7 were frequently affected compared with exons 5 or 8 (Fig 4). Among 96 point mutations, 86 mutations resulted in an amino acid change and four did not produce a change in amino acid type. Although codons for 18 different amino acids were affected by these mutations, the codon for arginine was most frequently affected. Codons 175, 248, and 273 were frequent targets and had 19% of all mutations (Fig 4). In six tumors, the point mutation in codons 196, 213, and 306 that code for arginine created a premature stop. These data indicate that the P53 mutation spectrum in B-NHL is not significantly different from that in other hematologic tumors.3 58 In addition, these data also indicate that in B-NHL, endogenous mutagenesis probably related to cytosine methylation is more frequent than mutations induced by exogenous mutagens.
The current study provides evidence for both recessive and dominant roles for P53 mutations in lymphoma. The majority of mutations (44%) were identified in tumors in which chromosome 17 was abnormal; in other words, they were associated with loss of the normal allele (loss of heterozygosity). These mutations might act as recessives. On the other hand, we also showed that P53 mutations were present in tumors in which both homologs of chromosome 17 appeared cytogenetically normal; in these tumors, mutant P53 presumably acts as a dominant negative mutation or with gain of function.
We examined the association between P53 mutation and cytogenetic structure of the tumor cells. Our data showed a positive association between P53 mutation and cytogenetic complexity of the tumor. Thus, all tumors with a mutation and successful karyotyping showed complex cytogenetic abnormalities. A similar association between P53 mutation and karyotypic complexity was identified in other hematologic malignancies9,59,60; however, it is not known whether these are causally related. The role of P53 in monitoring DNA replication and cell cycle46,47,50 61-63 suggests that P53 mutations could have been the cause of multiple chromosomal abnormalities, rather than the consequence; however, we have shown that the majority of tumors (80%) with complex cytogenetic abnormalities had no mutation in the exons examined. Therefore, unlike previous reports, we cannot conclude that P53 mutation induces instability in a genome leading subsequently to cytogenetic complexity. Our data indicate that mutation in P53 might occur concurrently with cytogenetic evolution in tumor cells.
A high level of p53 protein observed in tumors with mutated P53 has been associated with a longer half-life of mutated protein arising from conformational changes.64,65 In this study, we have presented data showing a lack of concordance between P53 mutation and p53 expression. There was a positive correlation between missense mutation and p53 expression and a negative correlation between non-missense mutation (premature stop codon, duplication, or deletion) and p53 expression. As reported previously for tumors of the lung, breast, and ovary,66 our data show that the majority of non-missense mutations prevent protein expression, and therefore, are most likely to be missed in protein studies.
We detected p53 in tumors without mutation in the exons examined and in many cases with clonal cytogenetic abnormality(ies), not affecting 17p. We cannot exclude the possibility of a mutation outside of exons 5 through 9 in these tumors as a cause of protein expression. However, previous studies have shown that these are infrequent in NHL67 and constitute less than 5% of mutations in general.68 These tumors, therefore, may show a mechanism of protein stabilization, independent of mutation, of a kind that has been observed with interaction with viral or other cellular proteins.46,69 Alternatively, in these tumors, our observations may represent a high level of constitutive expression associated with increased rate of cell division, as has been reported previously.67 Inactivation of p53 may lead to tumorigenesis. In fact, in liver cancer, it has been shown that functional inactivation of p53, but not mutations, leads to tumor development.70
The frequent finding of a mutation in P53 in advanced-stage disease or in tumors after histologic conversion is associated with a poor outcome.8,17,28,29,53,71-73 Indeed, two recent studies of mantle-cell lymphoma18,32 and one study of relapsed tumors53 have documented that P53 mutation and/or expression was associated with decreased survival, attributed to the development of new clones that are resistant to therapy. However, we identified P53 mutations at diagnosis in patients with DLC, which might suggest that, in these cases, P53 mutations were involved in early stages of the development of disease. In addition, we have also documented the clinical significance of mutation and/or expression. Among nonmutants, we found that complex cytogenetic abnormalities and p53 expression were associated with poor survival. Among different histologic groups, p53 expression was associated with poor survival in patients with FSCC and DLC tumors.
On the other hand, in our study, we observed that few patients with a mutation survived ≥5 years. This indicates that not all mutations confer an aggressive phenotype. Indeed, a recent study has shown that with some mutations in P53, the gene product behaves like the wild type and does not prevent tumor-suppressor activity.68
In conclusion, in this study, we identified several types of mutations, namely, missense and nonsense, that are structural in P53 in diagnostic and relapsed specimens of several histologic groups of NHL, but occur more frequently in tumors of DLC and mantle-cell histology. These mutations were detected in the presence or absence of cytogenetically abnormal 17p and were associated with complex cytogenetic abnormalities. There was no concordance between P53 mutation and p53 expression. In this regard, we have shown that 25% of tumors positive for p53 expression had mutations, whereas only 3% of tumors lacking expression had P53 mutations. Both mutation and protein expression have been found to negatively influence survival. Examination of P53 in these patients may therefore be useful in clinical management.
ACKNOWLEDGMENT
We are thankful to Barbara Napolitano of Division of Biostatistics for help with the statistical analysis of data.
Address reprint requests to Prasad R.K. Koduru, PhD, Cell Genetics Laboratory, North Shore University Hospital, 300 Community Dr, Manhasset, NY 11030.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal