Abstract
Mouse plasmacytomas share pathogenetic features in common with both multiple myeloma and Burkitt's lymphoma in humans. Susceptibility to plasmacytoma induction by intraperitoneal pristane in mice is controlled by multiple genes. At least two of these genes reside on mouse chromosome 4 in regions of the genome sharing linkage homology with human chromosomes 9p21, 1p32, and 1p36. A series of congenic strains recombinant for regions of mouse chromosome 4 in the vicinity of the Pctr2 predisposition locus were created and typed for their tumor susceptibility/resistance phenotypes. These strains were derived by introgressively backcrossing alleles from resistant DBA/2 mice onto the susceptible BALB/cAnPt background. Six resistant and two susceptible strains were allelotyped for 10 genes and 49 random DNA markers to identify the smallest region of overlap in the resistant strains. These studies have determined that the Pctr2 locus resides in either a 500-kb interval proximal to Nppa, or in a 1- to 2-centiMorgan (cM) interval distal to Nppa. In these congenic strain analyses, the Nppa and Fv1 loci, in addition to genes within about 1 cM of these loci, have been excluded as candidates for the Pctr2 locus. A relevant locus that may reside in this interval is Rep2; it is associated with the efficiency of repairing X-ray induced DNA damage sustained during the G2 phase of the mitotic cycle. The Pctr2 locus acts in a codominant fashion. F1 hybrids between resistant and susceptible congenic strains exhibit a reduced tumor incidence and a significant delay in the onset of tumorigenesis. Identification and eventual cloning of the Pctr2 locus may assist in the identification of genes involved in many types of cancer showing aberrations in human chromosome 1p36.
MOUSE PLASMACYTOMAS secrete Igs, predominantly of the IgA isotype,1 and greater than 95% of them carry translocations between the myc oncogene on chromosome 15 (Chr 15) and the Ig heavy chain locus on mouse Chr 12.2 As such, they represent a mouse model system for studying the pathogenesis of B-cell tumors such as Burkitt's lymphomas in humans, which harbor translocations involving the myelocytomatosis gene on human Chr 8 and the immunoglobulin heavy chain locus on human Chr 14.3 The model system involves the induction of plasmacytomas by the intraperitoneal injection of agents, such as pristane (2,6,10,14-tetramethylpentedecane), that induce a chronic inflammatory response. Genes affecting susceptibility/resistance phenotypes may affect this induction process. The chronic inflammatory response induced by pristane produces reactive oxygen intermediates that could induce DNA damage in nearby cells.4 Previous work has shown that BALB/c mice are relatively refractory in repairing double strand breaks in their DNA.5 6
To identify genes that predispose BALB/c mice to plasmacytoma development, two approaches have been used. First, backcross studies involving BALB/cAn and resistant DBA/2 mice were performed. Mock et al7 showed that mouse plasmacytomagenesis is a complex genetic trait with loci controlling the tumorigenic process linked to mouse Chr 1 near Fcgr2 and to the distal half of Chr 4. In these studies there were three distinct logarithm of the odds score peaks on Chr 4 near Ifna, Gt10, and Nppa (aliases Anf and Pnd) suggesting the possibility of more than one gene on Chr 4 controlling susceptibility/resistance patterns in backcross progeny. Resistance is almost completely dominant with a penetrance of 0.97 in F1 hybrids between BALB/c and DBA/2.7 Secondly, we constructed a series of BALB/c congenic strains that carried small segments of Chr 4 chromatin from the resistant strain DBA/2. At least two noncontiguous regions of Chr 4 contained plasmacytoma resistance genes, Pctr1 near Mtv13 and D4Lgm1, and Pctr2 near Nppa, in BALB/c.DBA/2 congenic strains of mice.8
The multigenic inheritance of a predisposition for plasmacytomagenesis seen in BALB/cAn mice precludes detailed fine-mapping of tumor susceptibility/resistance genes in simple genetic crosses. The goals of the current study were to refine the genetic interval surrounding the Pctr2 locus, evaluate whether the Fv1 locus was a likely candidate gene for Pctr2, and to determine the dominance characteristics of this resistance gene when isolated from the effects of the DBA/2 allele of Pctr1. To accomplish the task of narrowing genetic intervals of DBA/2 chromatin associated with resistance to tumor induction, a series of five new congenic strains were developed and tested for their tumor susceptibility phenotype associated with the Pctr2 locus. The results of these typings showed that Pctr2 was located in a 1- to 2.5-centiMorgan (cM) interval in the telomeric portion of mouse Chr 4. To determine the effects of the Pctr2 locus on tumor incidence and latency period, tumor inductions were performed in F1 hybrids between congenic strains that were homozygous D/D (C.D2-Fv1) and C/C (C.D2-IP3) at the Pctr2 locus. Both of these congenic strains were C/C at the Pctr1 locus in the mid-distal portion of Chr 4.
MATERIALS AND METHODS
Mice. C.D2-MIA mice were originally constructed by active selection of DBA/2 alleles/variants of Mtv13 (mammary tumor virus 13), D4Lgm1(1B4) (DNA segment, Chr 4; Lab of Genetics, Beverly A. Mock, NCI, Bethesda, MD) and Nppa (pronatrial naturietic factor) during introgressive backcrosses of DBA/2 to BALB/cAnPt mice8,9 (Fig 1). To produce recombinant strains across the 40 cM interval from D4Lgm1 to Nppa, C.D2-MIA (N11) mice were backcrossed to BALB/cAn. A number of recombinant strains were established as described.8 One of these, C.D2-Pnd7 (N12), carried DBA/2 alleles/variants of genes and markers near the Nppa locus at a position 77 cM from the centromere on mouse Chr 4. This strain was backcrossed to BALB/cAn to produce additional recombinants across smaller regions of Chr 4. In addition, another recombinant strain, C.D2-Pnd4 (N12), was derived from the C.D2-MIA (N11) mouse and further recombinants were also generated from this strain. In this study we have characterized C.D2-MIA (N10F15), C.D2-Pnd4(N12F14), C.D2-Pnd7 (N12F9), C.D2-Pnd4A (N15F5), C.D2-Pnd4C (N15F4), C.D2-Pnd7A (N15F3), C.D2-Pnd7B (N14F4), and C.D2-Fv1 (N19F13) mice for polymorphic variants of 59 DNA markers. In the set of experiments to test the dominance/recessive characteristics of the Pctr2 locus, C.D2-Idh1, Pep3 (C.D2-IP3) mice were used as susceptible controls; these mice carry DBA/2 alleles of markers in a 30-cM interval of mouse Chr 1 between Idh1 and Pep3.10,11 They are homozygous C/C at both Pctr1 and Pctr2 and have been found to be hypersusceptible to the induction of plasmacytomas by pristane.12 The mice were bred and maintained in conventional closed barrier conditions at PerImmune, Inc (Rockville, MD) under NCI contract N01-BC-21075. Mice were fed PMI 5001/5020 mouse chow (PMI Feeds, Inc, St. Louis, MO) and acidified water ad libitum.
Genealogy of the C.D2-Pnd strains. Backcross progeny between BALB/c and (BALB/c × DBA/2) F1 hybrids that contained DBA/2 alleles of distal Chr 4 markers were selected to create C.D2-MIA mice. At the N10 generation the C.D2-MIA mouse was heterozygous for all loci from Ifna to D4Mit254. Markers proximal to Ifna were homozygous C/C (BALB/c). At the N11 generation, recombinants near the Nppa locus were selected for further breeding to create the C.D2-Pnd4 and seven strains of mice. These mice were further bred to BALB/c to create additional recombinants in the distal Chr 4 region near Nppa to create the C.D2-Pnd4A,4C,7A and 7B strains.
Genealogy of the C.D2-Pnd strains. Backcross progeny between BALB/c and (BALB/c × DBA/2) F1 hybrids that contained DBA/2 alleles of distal Chr 4 markers were selected to create C.D2-MIA mice. At the N10 generation the C.D2-MIA mouse was heterozygous for all loci from Ifna to D4Mit254. Markers proximal to Ifna were homozygous C/C (BALB/c). At the N11 generation, recombinants near the Nppa locus were selected for further breeding to create the C.D2-Pnd4 and seven strains of mice. These mice were further bred to BALB/c to create additional recombinants in the distal Chr 4 region near Nppa to create the C.D2-Pnd4A,4C,7A and 7B strains.
Tumor inductions and diagnosis. Plasma cell tumors were induced in 6- to 10-week-old mice with three 0.5-mL, intraperitoneal injections of pristane (2,6,10,14-tetramethylpentadecane) on days 0, 60, and 120. A total of 40 mice per strain were used in each induction study. Thirty to 40 BALB/cAn mice were also used as controls in each induction experiment. BALB/c controls must be included for each experiment, because the incidence in BALB/c mice can vary from 30% to 65%8; the penetrance of the susceptible phenotype is under the influence of environmental factors. As such, it is always critical to compare a test strain for tumor incidence with BALB/c over the same time interval. Beginning at day 134, Wright's Giemsa-stained cytospin slides of ascites from peritoneal exudate cells were examined on a biweekly basis for the presence of atypical plasma cells. Mice were diagnosed as PCT-positive when accumulations of 10 or more atypical plasma cells were seen in two independent slides or when a single slide contained greater than 100 plasmacytoma cells. All studies were terminated at 300 days postpristane. Incidences of tumor-positive and -negative mice at day 300 were compared by 2 × 2 χ2 analyses.13
Fv1 phenotyping. Mice were biologically typed for Fv1 alleles using tissue cultures prepared from tail biopsies of weanling or young adult mice.14 In short, XC plaque assays were performed on duplicate tail cultures each infected with a dilution of standard N- (AKRL1) or B-tropic (WN1802B) murine leukemia viruses (MuLVs) chosen to give a clear distinction between BALB/c (Fv1b/b) and DBA/2 (Fv1n/n).15
Marker typing. DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blotting, and restriction fragment length polymorphism analyses were performed as described.7,16 DNA probes for Mtv13, D4Lgm1, D4Lgm3, Tnfr2, Nppa, and Pkcz-rs were described previously.8,9 Simple sequence length polymorphisms of simple sequence repeats were a modification of those previously described.17 Briefly, DNA (approximately 500 ng) was amplified by polymerase chain reaction (PCR) using a series of mouse map pairs for microsatellite sequences (DMIT's; Research Genetics, Huntsville, AL). PCRs were performed on either a PTC-100 (M.J. Research, Watertown, MA) or Omnigene (Hybaid, Franklin, MA) thermocycler using the following cycling conditions: 94°C for 1 minute, 55°C for 1 minute, 72°C for 30 seconds for 28 cycles, and a final extension at 72°C for 10 minutes. PCR products were visualized with ultraviolet light on either 6% to 16% polyacrylamide gels (Sequagel; National Diagnostics, Atlanta, GA) run on Bethesda Research Laboratories vertical gel rigs or 4% to 5% NuSieve 3:1 (FMC Bioproducts, Rockland, ME) horizontal agarose gels run on Mupid-2 (DNA Technologies, Inc, Rockville, MD) mini gel rigs and stained with ethidium bromide. As in previous studies,18 PCR assays for Xmv8, Xmv9, Xmv14, and Xmv44 were done with one common long terminal repeat (LTR)-specific oligonucleotide (SB1 5′CCT CAG ATT GAT TGA CTA CC3′) in combination with oligonucleotides specific for the 3′ sequences flanking the individual proviruses (Xmv8 SB2 5′GGC TGC TGG TTA TCT CTT GG3′; Xmv9 SB3 5′ACT GAC AGA GAT GGA GTT CC3′; Xmv14 SB4 5′TCC CGA CTC AAA TGC CAG C3′; Xmv44 SB5 5′CTG AAG AGA AAG CGC TGT GG3′). Mixed with the LTR primer, these oligonucleotides gave products of 297, 174, 259, and 227 bp in BALB/c mice. Cycling conditions for the Xmv loci were an initial denaturation at 94°C for 2 minutes followed by 32 cycles of 94°C for 1 minute, 55°C for 45 seconds, 72°C for 45 seconds, and a final extension at 72°C for 10 minutes. Primers used to detect a tetranucleotide repeat, referred to as D4Nimr1 (DNA segment, Chr 4; National Institute of Medical Research 1, Mill Hill, London, UK), located 30 kb from the 3′ side of the Fv1 locus were: PL40 5′AGG GAA TCT GAT GCC TTT GC3′ and PL41 5′TCA GTA AGT GTG CTT GCC TG3′. Cycling conditions for D4Nimr1 were 94°C for 1 minute, 62°C for 1 minute, and 72°C for 30 seconds for 32 cycles, followed by a final extension at 72°C for 10 minutes.
RESULTS AND DISCUSSION
Our earlier studies had shown that C.D2-MIA (N10F3) mice were resistant (4% incidence) to plasmacytomagenesis when compared with an incidence of 52% in BALB/cAnPt mice. These studies had also suggested that C.D2-Pnd7 (N12F9) mice were partially resistant; 5% of the mice had developed six or more plasmacytic foci in their mesenteries by day 144 compared with 29% in BALB/cAnPt.8 In the current study, we have examined later generations of these strains for full tumor incidences by 300 days postpristane. In addition, five new recombinant strains were derived from the original MIA and Pnd7 strains (Fig 1) and examined for their tumor susceptibility/resistance phenotypes as well as their allelic composition of markers on Chr 4.
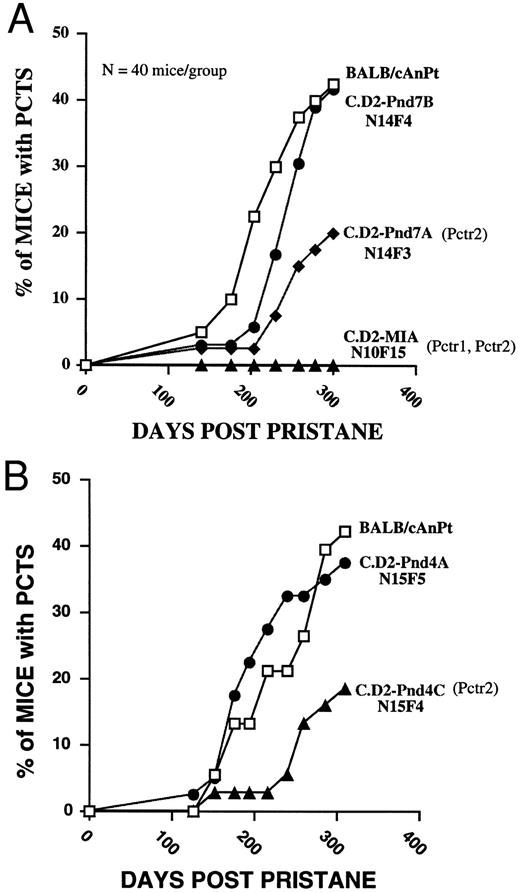
Tumor incidence patterns in congenic strains recombinant across the Pctr2 interval. The C.D2-MIA (N10F15) strain was solidly resistant (0% incidence compared with a final incidence of 42.5% in BALB/c mice at day 300) to plasma cell tumor formation over the course of 300 days (Fig 2A); the incidence in this strain was identical to that of DBA/2N mice from which its Chr 4 alleles of Mtv13, D4Lgm1, and Nppa were derived. This strain carries D/D alleles at both Pctr1 and Pctr2 and may also carry DBA/2 alleles of a third resistance gene located between 1 and 2. When the C.D2-Pnd7 (N12F9) mice were examined in the current study, we found that 23% of 43 mice had developed tumors by day 342 (day 300 data shown in Fig 2A) compared with 57% of 40 BALB/cAn mice. We also examined the tumor incidence patterns in later generations (N19F11) of the C.D2-Fv1 strain; the incidence of tumors in these mice was 14% (7 of 50) at day 300 compared with 37% in 30 BALB/c mice.
Tumor incidence patterns over the course of 300 days in BALB/cAn and the C.D2-Chr 4 congenic strains (N = 40 mice/strain). (A) Comparisons were made between BALB/c and C.D2-MIA, C.D2-Pnd7A, and C.D2-Pnd7B. (B) Comparisons were made between BALB/c and C.D2-Pnd4A, and C.D2-Pnd4C. The experiments illustrated in (A and B) represent two separate induction studies performed within 6 months of each other. See Table 1 for definition of the intervals of DBA/2 chromatin present in each of the C.D2-Chr 4 congenics. The C.D2-MIA mouse carries at least two resistance alleles at the Pctr1 and Pctr2 loci for plasmacytoma development. The Pnd7A and Pnd4C mice carry at least one resistance allele at the Pctr2 locus for plasmacytoma development. In contrast, the Pnd4A and Pnd7B mice were as susceptible as BALB/c and, therefore, were carrying susceptibility alleles at the Pctr2 locus.
Tumor incidence patterns over the course of 300 days in BALB/cAn and the C.D2-Chr 4 congenic strains (N = 40 mice/strain). (A) Comparisons were made between BALB/c and C.D2-MIA, C.D2-Pnd7A, and C.D2-Pnd7B. (B) Comparisons were made between BALB/c and C.D2-Pnd4A, and C.D2-Pnd4C. The experiments illustrated in (A and B) represent two separate induction studies performed within 6 months of each other. See Table 1 for definition of the intervals of DBA/2 chromatin present in each of the C.D2-Chr 4 congenics. The C.D2-MIA mouse carries at least two resistance alleles at the Pctr1 and Pctr2 loci for plasmacytoma development. The Pnd7A and Pnd4C mice carry at least one resistance allele at the Pctr2 locus for plasmacytoma development. In contrast, the Pnd4A and Pnd7B mice were as susceptible as BALB/c and, therefore, were carrying susceptibility alleles at the Pctr2 locus.
Two new congenic strains, C.D2Pnd7A and C.D2Pnd7B, which were recombinant across the Pnd7 interval (Table 1) of mouse Chr 4 differed in their tumor induction profiles (Fig 2A; day 300 χ2 = 5.2, P = .02). The tumor latencies and incidence patterns seen in C.D2-Pnd7B mice were like those seen in BALB/cAn susceptible mice (Fig 2A). In contrast, the C.D2-Pnd7A mice were partially resistant to plasmacytomagenesis with a final incidence of 20% at day 300 (Fig 2A) similar to that seen in the C.D2-Pnd7 progenitor strain (Fig 2A).
Allelotype Analysis of Resistant and Susceptible C.D2-Chr 4 Congenic Strains
| Marker* . | Pos. (cM) . | MIA . | Pnd4 . | Pnd4A N15F6 . | Pnd4C N15F7 . | Fv1 N19F11 . | Pnd7 N12F9 . | Pnd7A N15F7 . | Pnd7B N14F8 . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | N10F15 . | N12F14 . | . | . | . | . | . | . |
| % PCT + at 300d | 0% | 6% | 38% | 18% | 14% | 21% | 20% | 42% | |
| % PCT in BALB/c† | 43% | 37% | 42% | 42% | 37% | 57% | 43% | 42% | |
| Pctr2 Phenotype | R = D | PR = D | S = C | PR = D | PR = D | PR = D | PR = D | S = C | |
| Predicted Pctr1 | D | C | C | C | C | C | C | C | |
| D4Mit84 | 37.7 | C | C | C | C | C | C | C | C |
| Ifna, D4Mit27 | 42.6 | D | C | C | C | C | C | C | C |
| D4Mit15 | 43 | D | C | C | C | C | C | C | C |
| D4Mit153 | 45.5 | D | C | C | C | C | C | C | C |
| D4Mit28 | 46 | D | C | C | C | C | C | C | C |
| D4Mit119 | 49 | D | C | C | C | C | C | C | C |
| D4Mit187 | 49 | D | C | C | C | C | C | C | C |
| Mtv13, D4Mit31 | 49.5 | D | C | C | C | C | C | C | C |
| Tal2 (Scl) | 49.5 | D | C | C | C | C | C | C | C |
| D4Lgm1 | 50.6 | D | C | C | C | C | C | C | C |
| D4Rck41 | 51 | D | D | D | C | C | C | C | C |
| Ccnb1-rs10 | 50.7 | D | D | D | C | C | C | C | C |
| D4Mit52 | 54.9 | D | D | D | C | C | C | C | C |
| D4Mit57 | 56 | D | D | D | C | C | C | C | C |
| D4Mit122 | 56 | D | D | D | C | C | C | C | C |
| D4Mit37 | 56.5 | D | D | D | C | C | C | C | C |
| D4Mit148 | 66 | D | D | C | C | D | C | C | C |
| D4Mit284 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit312 | 69.8 | D | D | C | C | C | C | C | C |
| Gt10 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit65 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit32 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit252 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit48 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit13 | 71 | D | D | C | C | C | C | C | C |
| D4Mit126 | 71 | D | D | C | C | C | C | C | C |
| D4Mit128 | 71 | D | D | C | C | C | C | C | C |
| D4Mit343 | 71 | D | D | C | C | C | C | C | C |
| D4Mit341 | 71 | D | D | C | D | C | C | C | C |
| D4Lgm3 | 75.3 | D | D | C | D | C | C | C | C |
| Tnfr2 (D4Mit233) | 75.5 | D | D | C | D | D | C | C | C |
| D4Mit160 | 76 | D | D | C | D | D | C | C | C |
| D4Mit340 | 76 | D | D | C | D | D | C | C | C |
| D4Mit49 | 76 | D | D | C | D | D | C | C | C |
| D4Mit259 | 76 | D | D | C | D | D | C | C | C |
| D4Mit285 | 76 | D | D | C | D | D | C | C | C |
| Xmv8 | 76.2 | D | D | C | D | D | C | C | C |
| Xmv44 | 76.4 | D | D | C | D | D | D | D | D |
| Xmv14 | 76.5 | D | D | C | D | D | D | D | D |
| Xmv9 | 76.5 | D | D | C | D | D | D | D | D |
| D4Rck128 | 76.5 | D | D | C | D | D | D | ||
| Nppa (Pnd) | 76.5 | D | D | C | D | D | D | D | D |
| Fv1 | 76.5 | D | D | C | D | D | D | D | D |
| D4Nimr1 | 76.5 | D | D | C | D | D | D | D | D |
| D4Mit310 | 77 | D | D | C | D | D | D | D | C |
| D4Mit129 | 77 | D | D | C | D | D | D | D | C |
| D4Mit190 | 79 | C | D | C | D | D | D | D | C |
| D4Mit33 | 79 | C | D | C | D | D | D | D | C |
| D4Mit226 | 79 | C | D | C | D | D | D | D | C |
| D4Mit42 | 81 | C | D | C | D | D | D | D | C |
| D4Mit180 | 81 | C | D | C | D | D | D | D | C |
| D4Mit131 | 81.2 | C | D | C | D | D | D | D | C |
| D4Mit253 | 81.2 | C | D | C | D | D | D | D | C |
| D4Mit313 | 81.5 | C | D | C | D | D | D | C | C |
| D4Mit254 | 81.7 | C | D | C | D | D | D | C | C |
| Pkcz-rs | 81.9 | C | D | C | D | C | D | C | C |
| D4Smh6b | 82 | C | D | C | D | C | D | C | C |
| Marker* . | Pos. (cM) . | MIA . | Pnd4 . | Pnd4A N15F6 . | Pnd4C N15F7 . | Fv1 N19F11 . | Pnd7 N12F9 . | Pnd7A N15F7 . | Pnd7B N14F8 . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | N10F15 . | N12F14 . | . | . | . | . | . | . |
| % PCT + at 300d | 0% | 6% | 38% | 18% | 14% | 21% | 20% | 42% | |
| % PCT in BALB/c† | 43% | 37% | 42% | 42% | 37% | 57% | 43% | 42% | |
| Pctr2 Phenotype | R = D | PR = D | S = C | PR = D | PR = D | PR = D | PR = D | S = C | |
| Predicted Pctr1 | D | C | C | C | C | C | C | C | |
| D4Mit84 | 37.7 | C | C | C | C | C | C | C | C |
| Ifna, D4Mit27 | 42.6 | D | C | C | C | C | C | C | C |
| D4Mit15 | 43 | D | C | C | C | C | C | C | C |
| D4Mit153 | 45.5 | D | C | C | C | C | C | C | C |
| D4Mit28 | 46 | D | C | C | C | C | C | C | C |
| D4Mit119 | 49 | D | C | C | C | C | C | C | C |
| D4Mit187 | 49 | D | C | C | C | C | C | C | C |
| Mtv13, D4Mit31 | 49.5 | D | C | C | C | C | C | C | C |
| Tal2 (Scl) | 49.5 | D | C | C | C | C | C | C | C |
| D4Lgm1 | 50.6 | D | C | C | C | C | C | C | C |
| D4Rck41 | 51 | D | D | D | C | C | C | C | C |
| Ccnb1-rs10 | 50.7 | D | D | D | C | C | C | C | C |
| D4Mit52 | 54.9 | D | D | D | C | C | C | C | C |
| D4Mit57 | 56 | D | D | D | C | C | C | C | C |
| D4Mit122 | 56 | D | D | D | C | C | C | C | C |
| D4Mit37 | 56.5 | D | D | D | C | C | C | C | C |
| D4Mit148 | 66 | D | D | C | C | D | C | C | C |
| D4Mit284 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit312 | 69.8 | D | D | C | C | C | C | C | C |
| Gt10 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit65 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit32 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit252 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit48 | 69.8 | D | D | C | C | C | C | C | C |
| D4Mit13 | 71 | D | D | C | C | C | C | C | C |
| D4Mit126 | 71 | D | D | C | C | C | C | C | C |
| D4Mit128 | 71 | D | D | C | C | C | C | C | C |
| D4Mit343 | 71 | D | D | C | C | C | C | C | C |
| D4Mit341 | 71 | D | D | C | D | C | C | C | C |
| D4Lgm3 | 75.3 | D | D | C | D | C | C | C | C |
| Tnfr2 (D4Mit233) | 75.5 | D | D | C | D | D | C | C | C |
| D4Mit160 | 76 | D | D | C | D | D | C | C | C |
| D4Mit340 | 76 | D | D | C | D | D | C | C | C |
| D4Mit49 | 76 | D | D | C | D | D | C | C | C |
| D4Mit259 | 76 | D | D | C | D | D | C | C | C |
| D4Mit285 | 76 | D | D | C | D | D | C | C | C |
| Xmv8 | 76.2 | D | D | C | D | D | C | C | C |
| Xmv44 | 76.4 | D | D | C | D | D | D | D | D |
| Xmv14 | 76.5 | D | D | C | D | D | D | D | D |
| Xmv9 | 76.5 | D | D | C | D | D | D | D | D |
| D4Rck128 | 76.5 | D | D | C | D | D | D | ||
| Nppa (Pnd) | 76.5 | D | D | C | D | D | D | D | D |
| Fv1 | 76.5 | D | D | C | D | D | D | D | D |
| D4Nimr1 | 76.5 | D | D | C | D | D | D | D | D |
| D4Mit310 | 77 | D | D | C | D | D | D | D | C |
| D4Mit129 | 77 | D | D | C | D | D | D | D | C |
| D4Mit190 | 79 | C | D | C | D | D | D | D | C |
| D4Mit33 | 79 | C | D | C | D | D | D | D | C |
| D4Mit226 | 79 | C | D | C | D | D | D | D | C |
| D4Mit42 | 81 | C | D | C | D | D | D | D | C |
| D4Mit180 | 81 | C | D | C | D | D | D | D | C |
| D4Mit131 | 81.2 | C | D | C | D | D | D | D | C |
| D4Mit253 | 81.2 | C | D | C | D | D | D | D | C |
| D4Mit313 | 81.5 | C | D | C | D | D | D | C | C |
| D4Mit254 | 81.7 | C | D | C | D | D | D | C | C |
| Pkcz-rs | 81.9 | C | D | C | D | C | D | C | C |
| D4Smh6b | 82 | C | D | C | D | C | D | C | C |
Shaded areas designate possible locations for the Pctr2 locus.
Abbreviations: Pos, consensus map position in cM from the centromere; N, backcross generations of the congenic; F, filial generations of the congenic; % PCT, percent of PCT-positive mice at day 300; R, resistant; D, DBA/2N; PR, partially resistant; S, susceptible; C, BALB/cAn.
Marker refers to the Chr 4 locus examined.
% PCT in BALB/c refers to the incidence of plasmacytomagenesis in 30 or 40 BALB/c controls tested simultaneously with the designated congenic strain.
An additional three new congenics were also compared in tumor induction studies. The C.D2-Pnd4 strain was derived from N11 C.D2-MIA mice (Fig 1) and was found to be resistant to tumor formation with a final incidence of 6% at day 300 compared with 37% in BALB/c mice (data not shown). When the Pnd4 interval was further partitioned by creating two new recombinant strains, C.D2-Pnd4A and C.D2-Pnd4C, it was found that Pnd4A mice were highly susceptible like the BALB/cAn control mice (Fig 2B). In contrast, C.D2-Pnd4C mice were partially resistant with an incidence of 15% by day 300. The percent of mice with plasmacytomas at day 300 was higher in BALB/cAn than Pnd4C (χ2 = 5.05, P = .02). Similarly, the percent of mice in the Pnd4A strain with plasma cell tumors was also higher than that of the Pnd4C strain at day 300. These data imply that the Pnd4A strain is C/C at both Pctr1 and Pctr2.
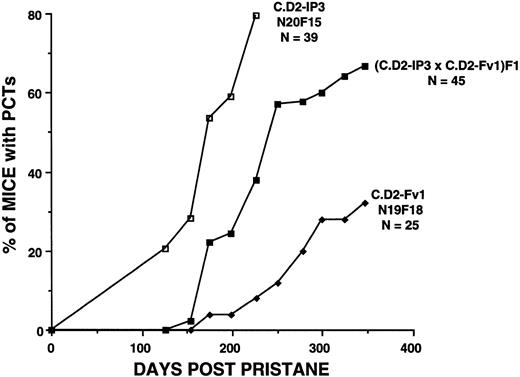
F1 hybrids between resistant and susceptible congenics. To evaluate the dominance characteristics of the Pctr2 locus, the partially resistant C.D2-Fv1 congenic was crossed to the hypersusceptible congenic strain, C.D2-Idh1, Pep3. F1 hybrids between these two strains of mice exhibited an intermediate phenotype with respect to tumor incidence patterns over the course of 347 days (Fig 3). At day 226, incidence patterns of the F1 hybrids were compared with those of the two parental strains. Studies with the IP3 mice were terminated at day 226 because of significant morbidity and mortality in these mice. Fewer F1 hybrid mice (38%) had tumors than C.D2-IP3 mice (80%; χ2 = 11.3, P = .0007), and more F1 hybrid mice had tumors than the C.D2-Fv1 mice (8%; χ2 = 7.2, P = .007).
Tumor incidence patterns over the course of 350 days in F1 hybrids between partially resistant C.D2-Fv1 and hypersusceptible C.D2-Idh1, Pep3 (C.D2-IP3) mice. Studies were terminated in the C.D2-IP3 mice at day 226 because of high morbidity and mortality in this strain. Resistance was semidominant in the F1 hybrids between Fv1 and IP3 mice.
Tumor incidence patterns over the course of 350 days in F1 hybrids between partially resistant C.D2-Fv1 and hypersusceptible C.D2-Idh1, Pep3 (C.D2-IP3) mice. Studies were terminated in the C.D2-IP3 mice at day 226 because of high morbidity and mortality in this strain. Resistance was semidominant in the F1 hybrids between Fv1 and IP3 mice.
The F1 hybrid mice have inherited one resistance allele of DBA/2 origin and one susceptibility allele of BALB/c origin at the Pctr2 locus. Resistance and susceptibility alleles were semidominant or codominant in genetic crosses between C.D2-Fv1 and C.D2-IP3 mice. This is in contrast to the 0% to 3% tumor incidence and long latency period (400 days) seen in F1 hybrids between BALB/c and DBA/2.7 19 CxDF1 hybrids are carrying at least one resistance allele at each of the three or more tumor susceptibility loci involved in the genetic control of plasmacytomagenesis in BALB/c mice. Phenotypic effects of the Pctr2 locus were evident in delaying the onset of tumorigenesis and in the final tumor incidences by day 347 (Fig 3).
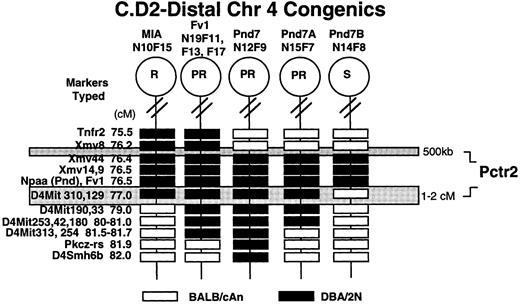
Polymorphic variants of genes/markers in the distal Chr 4 congenics. A total of eight C.D2 congenic strains of mice were allelotyped for a series of 10 genes and 49 microsatellite markers on the distal half of mouse Chr 4 to delineate the ends of each selected interval. The C.D2-MIA(N10F15) strain has 40 cM of DBA/2 chromatin from Ifna to D4Mit129 and, as such, contains DBA/2 alleles at both Pctr1 and Pctr2 loci7,8 (Table 1, Figs 1 and 4). Earlier generations (N10F3) of the C.D2-MIA mice were carrying DBA/2 alleles of genes across a larger interval from Ifna to D4Smh6b.8 Loci between D4Mit129 and D4Smh6b were still segregating in the earlier filial generations and became fixed as homozygous “C” by N10F15. The C.D2-Pnd4 and C.D2-Pnd7 strains were derived from the MIA strain at the N11 backcross generation. Because these mice were selected only for the Nppa locus, the telomeric end of the DBA/2 chromatin varied in length. At N10F15, the length of DBA/2 chromatin was shorter and did not extend beyond the D4Mit129 marker. The Pnd4 strain contains a 32-cM interval of DBA/2 chromatin from D4Lgm1 to D4Smh6b whereas the Pnd7 strain contains only 5 cM of DBA/2 chromatin tightly linked to the Nppa locus (Table 1, Figs 1 and 4). In addition, further backcrosses of the C.D2-Fv1 strain to BALB/cAn resulted in a partially PCT-resistant strain with a 7-cM interval of DBA/2 alleles/variants, overlapping the Pnd7 interval, in the telomeric portion of the chromosome.
Haplotype analyses of the C.D2-Chr 4 congenics showing the smallest region of overlap among the resistant strains to delineate the interval surrounding the Pctr2 locus. The markers typed and their map positions in centiMorgans (cM) are indicated to the left. The C.D2-MIA strain at N10F15 was solidly resistant (0% incidence) and is likely to be homozygous for two (Pctr1 and Pctr2 ) or more resistance genes from DBA/2, which influence plasmacytomagenesis. The C.D2-Fv1 and C.D2-Pnd7 strains have inherited only one resistance locus (Pctr2 ) from DBA/2. Both C.D2-Pnd7A and Pnd7B were capable of restricting the replication of B-tropic viruses, and both had inherited the same allele of Fv1, yet the two strains differed in their tumor induction profiles (see Fig 2A). The Pnd7B strain was susceptible to plasmacytomagenesis; therefore, this interval does not contain the Pctr2 locus. Open boxes refer to the BALB/cAn allele and darkened boxes, the DBA/2 allele.
Haplotype analyses of the C.D2-Chr 4 congenics showing the smallest region of overlap among the resistant strains to delineate the interval surrounding the Pctr2 locus. The markers typed and their map positions in centiMorgans (cM) are indicated to the left. The C.D2-MIA strain at N10F15 was solidly resistant (0% incidence) and is likely to be homozygous for two (Pctr1 and Pctr2 ) or more resistance genes from DBA/2, which influence plasmacytomagenesis. The C.D2-Fv1 and C.D2-Pnd7 strains have inherited only one resistance locus (Pctr2 ) from DBA/2. Both C.D2-Pnd7A and Pnd7B were capable of restricting the replication of B-tropic viruses, and both had inherited the same allele of Fv1, yet the two strains differed in their tumor induction profiles (see Fig 2A). The Pnd7B strain was susceptible to plasmacytomagenesis; therefore, this interval does not contain the Pctr2 locus. Open boxes refer to the BALB/cAn allele and darkened boxes, the DBA/2 allele.
Four recombinant strains were generated by crossing C.D2-Pnd4 and C.D2-Pnd7 mice back to BALB/cAn and selecting progeny that contained different intervals of Chr 4 derived from DBA/2 (Fig 1). The two strains, C.D2-Pnd4A and C.D2-Pnd4C, derived from the Pnd4 mice, contained noncontiguous, nonoverlapping intervals of Chr 4 separated by 19 cM (Table 1, Fig 4). The susceptible Pnd4A mouse contained a 6-cM interval between D4Lgm1 and D4Mit37 derived from DBA/2. In contrast, the resistant Pnd4C strain had a 12-cM interval stretching from D4Mit341 to D4Smh6b derived from DBA/2. As may be seen in Table 1, this segment overlaps the DBA/2 segments found in the C.D2-Fv1, C.D2-Pnd7, and C.D2-Pnd7A strains, whereas the DBA/2 segment in the Pnd4A strain did not.
The two recombinant strains, C.D2-Pnd7A and C.D2-Pnd7B, covering the telomeric portion of the chromosome made from the Pnd7 mouse were more informative (Fig 1). The Pnd7A strain contains DBA/2 alleles of markers across 5 cM from Xmv44 to D4Mit180. The susceptible Pnd7B strain contains an interval of Chr 4 directly overlapping a 1-cM segment within the resistant Pnd7A strain (Fig 4). A combination of congenic strain and yeast artificial chromosome (YAC) analyses (Best S, PhD Thesis, The positional cloning of Fv-1: a mouse retroviral restriction gene. University of London, 1994) have determined the following gene order for the Xmv loci: Xmv8 — Xmv44 — Xmv14/Xmv9. Earlier studies had shown that Xmv9 mapped distal to the other three Xmv loci.18
Fv1 is not a candidate for the Pctr2 locus. In this and previous studies, the BALB/c.DBA/2-Fv1n/n congenic strain was found to be partially resistant to PCT induction suggesting that the Pctr2 resistance gene is located in the vicinity of Fv1.8,20 This locus is responsible for controlling retroviral replication by N- or B-tropic ecotropic MuLVs21 and could, therefore, be important in controlling the development of virally induced tumors.22
When the PCT-resistant Pnd7A and PCT-susceptible Pnd7B mice were examined for their alleles at the Fv1 locus by phenotypic assays, it was found that both Pnd7A and Pnd7B mice were capable of restricting the viral replication of B-tropic viruses like DBA/2 mice. The mean titers in pfu log10 /0.2 mL for AKRL1 N-tropic virus in C.D2-Pnd7A, C.D2-Pnd7B, DBA/2N, and BALB/cAn were 4.9, 4.9, 4.5, and 2.8, respectively. In contrast, the titers for the WN1802 B-tropic virus were the following: Pnd7A (3.0), Pnd7B (3.0), DBA/2 (2.6), and BALB/cAn (5.0). When the Fv1 locus was cloned,23 the mice were also tested by PCR for their Fv1 alleles using the D4Nimr1 marker located just 30 kb on the 3′ side of Fv1 and, as expected from the in vivo studies, both congenic strains carried the DBA/2 allele. Thus, the 1-cM interval of DBA/2 chromatin found in the susceptible C.D2-Pnd7B strain contains at least three known genes, Nppa, Nppb (pronatrial naturietic factor, α and β), and Fv1, which are, therefore, excluded as candidates for the Pctr2 locus.
Definition of the interval surrounding the Pctr2 locus. The allelotypic analyses (Table 1) have defined an interval of mouse Chr 4 (Fig 4) containing the Pctr2 locus involved in controlling the resistance of DBA/2 mice to plasma cell tumorigenesis. The smallest region of overlap among the resistant and susceptible strains suggested that the Pctr2 gene resides in either of two locations: (1) within the interval between Xmv8 and 44 or (2) in the 1- to 2-cM interval between Nppa and D4Mit190. The distance between Xmv8 and Xmv44 has been determined to be ≤500 kb as a single YAC clone of this size has been identified, which harbors both loci (Best S, PhD Thesis). Current studies are focused on refining the genetic map in this interval by both backcross and congenic strain analyses. We have generated C.D2 congenics that contain DBA/2 alleles across either the 500 kb interval just proximal or the 1- to 2.5-cM region just distal to the Nppa, Nppb, Fv1 exclusion cluster and are in the process of making the strains homozygous for determining their tumor phenotype. Once we have established that the Pctr2 locus resides within an interval of 1 cM or less, we will construct a physical map of the region and search for B-cell–specific transcripts within the critical region.
Possible candidate genes in the telomeric portion of Chr 4 include Cd30, Pgd, Ox40, Cdc2l1, Eno1, and Gpd1. Their exact map positions relative to D4Mit310 and D4Mit129 require further evaluation. Of note, phenotypes associated with Rep2 (repair of chromatin damage 2),5,6Wld (Wallerian nerve degeneration),24 and Ssm1 (strain specific modifier of methylation)25 have also been localized to the subtelomeric portion of mouse Chr 4. The Rep2 phenotype is of special interest because C.D2-Fv1 mice were found to repair X-ray–induced double-strand breaks in their DNA more efficiently than BALB/c.5,6 Many of the developmental steps in pristane-induced plasmacytomagenesis take place in chronic inflammatory tissues induced by pristane. In this environment, B cells and plasma cells are exposed to a microenvironment that generates DNA damaging reactive oxygen intermediates.4 An inefficiency of DNA repair in BALB/c B or plasma cells could lead to an accumulation of mutational changes that may play a role in plasmacytomagenesis.
The Xmv44 to D4Mit310 region of mouse Chr 4 shares a large degree of synteny with human chromosome 1p36.26 Cytogenetic changes in Chr 1 are frequent in many forms of human cancer.27 In neuroblastoma, loss of heterozygosity studies have pinpointed 1p36.2 to 1p36.3 near natriuretic peptide precursor A as the region most consistently involved.28-30 Additional deletions and translocations involving this region of human Chr 1 have also been seen in colorectal cancer, breast cancer, melanoma, multiple endocrine neoplasia type II, hepatocellular carcinoma, pancreatic adenocarcinoma, cervical carcinoma, and myelodysplastic syndromes (reviewed in Schwab et al27 ). We are in the process of analyzing tumors induced in F1 hybrid mice to determine whether loss of heterozygosity, a feature frequently associated with the loss of a tumor suppressor allele, may occur in the interval surrounding the Pctr2 locus. The fine-mapping and eventual isolation of this plasmacytoma susceptibility locus may lead to the identification of a gene commonly altered in many forms of cancer.
ACKNOWLEDGMENT
The authors thank Jonathan Stoye and an anonymous reviewer for their valuable insights and comments on the original manuscript. We also wish to thank the following persons for their expert technical assistance in the production and genotyping of the C.D2 strains of mice: Wendy DuBois, David Bernard, Simon Leath, Danielle Holiday, Marianne Henderson, Jennifer Dosik, Jennifer Spence, and Beth Wieser.
Address reprint requests to Beverly A. Mock, PhD, National Cancer Institute, National Institutes of Health, 37 Convent Dr, MSC 4255, Building 37, Rm 2B08, Bethesda, MD 20892.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal