Abstract
Congenital dyserythropoietic anemia type II (CDA-II) is the most common form of inherited dyserythropoiesis. Previous studies have shown that the anion transporter (band 3) is narrower and it migrates faster on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); this aspect was related to insufficient glycosylation. Biochemical data support the hypothesis that this disease is due to a deficiency of N-acetylglucosaminyltransferase II (GnT II) or α-Mannosidase II (α-Man II), which represent the key to glycosylation. In addition, a third candidate gene is α-Man IIx, which shows a strong homology with α-Man II. The knowledge of the chromosomal localization of these putative genes allowed us to perform a linkage study using three sets of microsatellite markers flanking the candidate genes. Six families with two or more affected children were enrolled in this study. The data obtained exclude linkage to all three candidate genes. In consideration of the biochemical data (reduction of enzymatic activity) of the same enzymes, our results suggest the hypothesis that a defect in an unknown transcriptional factor is involved in CDA-II.
CONGENITAL DYSERYTHROPOIETIC anemia type II (CDA-II) is an autosomal recessive disease1 that represents the most common form of CDA. Like other forms, it is characterized by a mild to moderate lifelong anemia, ineffective erythropoiesis, and morphologic abnormalities of mature red blood cells (RBC) and their precursors.2 Splenomegaly and jaundice are the most evident clinical signs.3 4
The erythrocytes from patients with CDA-II showed an increased agglutinability and lysis to anti-i and anti-I sera.2 Ii are carbohydrate antigens present on the erythrocyte surface and they are determined by branched (I) and linear (i) poly-N-acetyllactosamines.5 Moreover, they are lysed when incubated in acidified serum by an alloantibody present in normal sera (Ham's test).2 6 Consequently, CDA-II is also called HEMPAS (hereditary erythroblastic multinuclearity associated with a positive acidified serum test).
Diagnosis was, for a long time, achieved by bone marrow examination. It shows 5 to 10 times more erythroblasts than normal (erythroid hyperplasia); early erythroblasts are relatively normal, but about 10% to 40% of more mature erythroblasts are binucleated or multinucleated.2-4 Extensive morphological anomalies of CDA-II erythroblastic cells were observed using electron microscopy, with the most significant being the presence of the so-called double membrane, which is observable in mature RBCs as well.7-9
More recently, Alloisio et al10 described the presence of three minor proteins (glucose-regulated protein [GRP] 78, calreticulin, and protein disulfide isomerase [PDI]) in the RBC membranes of CDA-II patients. All of these are major constitutive proteins of the endoplasmic reticulum lumen. This presence strongly supports the hypothesis that the second membrane must arise from the endoplasmic reticulum.10
Membrane protein analysis by means of polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) showed that the anion transporter (band 3) is narrower and migrates faster. This aspect was related to insufficient incorporation of lactosaminoglycans.11,12 Hence, the CDA-II carbohydrate structure analysis of membrane and soluble glycoproteins indicates that CDA-II is associated with defects in the biosynthesis of complex N-linked oligosaccharides.13
The presence in CDA-II patients of tri-mannosyl and penta-mannosyl hybrid-type oligosaccharides has led to the suggestion that the glycosylation defect is the result of a deficiency of either N-acetylglucosaminyltransferase II (GnT-II) or α-Mannosidase II (α-Man II) activity,14,15 key enzymes catalyzing synthesis of N-glycans of glycoproteins.4 Their corresponding genes are probably widely expressed, as shown by the presence of serum glycoproteins with incompletely processed carbohydrates circulating in the plasma of HEMPAS patients.16
At present it is not clear whether CDA-II is genetically heterogeneous or homogenous, but it appears as a heterogeneous enzymatic defect of glycosylation enzymes.14-17 Cloning and mapping of the putative genes makes possible molecular analysis; however, no linkage studies with this disorder have previously been performed. The knowledge of the localization of candidate genes allowed for the study of linkage in families with multiple affected children by using highly polymorphic markers (microsatellites).
To establish the specific role of these genes in the CDA-II, we performed linkage analysis of candidate genes (GnT-II, α-Man II, and α-Man IIx) with the clinical phenotype in six families with at least two affected siblings. Our results showed for the first time that these putative genes are not primarily related to the disease.
MATERIALS AND METHODS
A linkage analysis study was performed to examine the possible involvement of three candidate genes in determining CDA-II. For this purpose, highly informative markers located at the same chromosomal region of the candidate genes were used. Candidate genes and their chromosomal location were α-Man II, which is mapped to chromosome 5 (5q2.1-2.2); α-Man IIx isozyme mapped to chromosome 15q2518,19; and human Gnt II, which is mapped to the long arm of chromosome 14 (14q21).20 To speed up the analysis, four unrelated families containing at least two CDA II individuals and two kindreds with three affected patients were selected and included in the study. No consanguineity was established in any case. Thus, a total of 14 affected individuals were included in the study. The main clinical and biochemical aspects regarding six patients belonging to five different kindreds have been reported elsewhere (see subjects CE, DO, GE, RO II.1, RO II.2, and VA in Alloisio et al10 ). The other eight patients were found to have anemia of variable degrees, jaundice, and mild splenomegaly. Bone marrow observation showed the presence of erythroid hyperplasia with binuclearity and multinuclearity in approximately 10% to 40% of more mature erythroblasts. Erythrocyte protein membrane analysis was performed using SDS-PAGE.10 Western blotting was performed after SDS-PAGE. A monoclonal antibody against a synthetic peptide of rat glucose-regulated protein (GRP)-78 (Stress Gen Biotechnologies Corp, Victoria, British Columbia, Canada) and a polyclonal rabbit antibody against bovine protein disulfide isomerase (PDI; Stress Gen Biotechnologies Corp) were used as previously reported.10
Pairwise Iod Scores Between CDA-II and Chromosome Markers
| . | Recombination Frequencies . | ||||||
|---|---|---|---|---|---|---|---|
| . | 0 . | .01 . | .05 . | .1 . | .2 . | .3 . | .4 . |
| Chromosome 15 (α-Man II) | |||||||
| D15S205 | ∞ | −3.30 | −1.39 | −0.68 | −0.18 | −0.03 | −0.01 |
| D15S152 | ∞ | −4.14 | −1.57 | −0.66 | −0.05 | −0.06 | −0.03 |
| Chromosome 5 (α-Man II x) | |||||||
| D5S433 | ∞ | −5.25 | −2.06 | −0.92 | −0.17 | 0.01 | 0.02 |
| D5S2051 | ∞ | −7.54 | −3.55 | −2.02 | −0.76 | −0.26 | −0.05 |
| D5S421 | ∞ | −2.33 | −0.51 | 0.04 | 0.26 | 0.17 | 0.05 |
| Chromosome 14 (GnT-II) | |||||||
| D14S978 | ∞ | −7.24 | −3.27 | −1.77 | −0.58 | −0.16 | −0.02 |
| D14S281 | ∞ | −8.12 | −4.07 | −2.45 | −1.03 | −0.39 | −0.09 |
| D14S1064 | ∞ | −4.44 | −1.85 | −0.91 | −0.24 | −0.04 | −0.03 |
| . | Recombination Frequencies . | ||||||
|---|---|---|---|---|---|---|---|
| . | 0 . | .01 . | .05 . | .1 . | .2 . | .3 . | .4 . |
| Chromosome 15 (α-Man II) | |||||||
| D15S205 | ∞ | −3.30 | −1.39 | −0.68 | −0.18 | −0.03 | −0.01 |
| D15S152 | ∞ | −4.14 | −1.57 | −0.66 | −0.05 | −0.06 | −0.03 |
| Chromosome 5 (α-Man II x) | |||||||
| D5S433 | ∞ | −5.25 | −2.06 | −0.92 | −0.17 | 0.01 | 0.02 |
| D5S2051 | ∞ | −7.54 | −3.55 | −2.02 | −0.76 | −0.26 | −0.05 |
| D5S421 | ∞ | −2.33 | −0.51 | 0.04 | 0.26 | 0.17 | 0.05 |
| Chromosome 14 (GnT-II) | |||||||
| D14S978 | ∞ | −7.24 | −3.27 | −1.77 | −0.58 | −0.16 | −0.02 |
| D14S281 | ∞ | −8.12 | −4.07 | −2.45 | −1.03 | −0.39 | −0.09 |
| D14S1064 | ∞ | −4.44 | −1.85 | −0.91 | −0.24 | −0.04 | −0.03 |
High molecular weight DNA was extracted from peripheral blood leukocytes using an automatic DNA extractor (ABI DNA PURE) according to the protocols of the manufacturers (Perkin Elmer, Foster City, CA). Fluorochrome-labeled primers were prepared by chemically attaching a fluorescent dye to the 5′-end of each forward oligonucleotide primer employing the fluorescent amidite reagent, 6-FAM amidite. Names and primer sequences for detecting D5S433, D5S2051, D5S421, D14S281, D14S978, D14S1064, D15S205, and D15S152 alleles were previously described.21,22 Polymerase chain reactions (PCR) were prepared using a BIOMEK 1000 robot station (Beckman Instrumentation, Fullerton, CA) and performed on an automated Thermal cycler (Perkin Elmer, Norwalk, CT) using conditions previously described for each microsatellite marker. After PCR amplification, 5 μL of each sample was precipitated by the addition of 400 μL of 100% ethanol. After centrifugation, dried pellets were resuspended in 5 μL of deionized water. Multiplexing was performed before loading the gel, mixing 1 μL of each sample before ethanol precipitation. Acrylamide gel electrophoresis was performed in 8 mol/L urea containing 1× Tris-Borate-EDTA (TBE), with the length and acrylamide percentage changing depending on the resolution required. For all gels, the acrylamide/bisacrylamide (N,N′-methylene-bis-acrylamide) ratio was 19:1 (wt/wt) for a cross-linking concentration of 5%. Reactions were analyzed either on 4.75% or 6% (wt/vol) gels with 24-cm well-to-read distances. Data were collected using the GeneScan Data Collection program version 1.1 and analyzed using the GeneScan Data Analysis software version 1.1, 1.2, or 1.2.1b1 (Applied Biosystems Inc, Foster City, CA). Correct allele size was assigned using the Genotyper software. Linkage was assessed by analysis of allele segregation of the eight different polymorphic markers (D5S433, D5S2051, D5S421, D14S978, D14S281, D14S1064, D15S205, and D15S152). Pairwise linkage analysis was performed using MLINK and ILINK from the FASTLINK program package (version 2.2).23 Gene frequency was based on disease prevalence (Θ = .001), and full penetrance was also assumed. Calculations were performed at recombination fractions (Θ) of 0, 0.05, 0.10, 0.20, 0.30, and 0.40 between each marker and the disease locus. The HOMOG program was used to test for nonallelic heterogeneity.
RESULTS
The clinical features as well as the characteristic morphological abnormalities of the bone marrow allowed us to perform CDA II diagnosis in all of the patients investigated. Diagnosis was further supported by the presence on SDS-PAGE of a band 3 with a narrower aspect and a faster migration. The presence of GRP 78 and PDI in the erythrocyte membrane, in agreement with findings observed in the patients previously reported,10 was shown on Western blotting (not shown).
Results of linkage analysis performed between CDA-II phenotype in the six index families versus specific chromosomal regions containing the candidate genes are reported in Table 1.
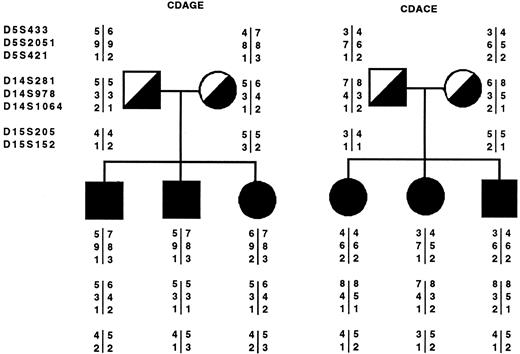
Microsatellite markers flanking the GnT-II, α-Man II, and α-Man IIx genes always gave negative lod scores at recombination frequencies of 0.0, 0.01, and 0.05. Only marker D5S421 showed a positive but not significant lod score of 0.04 at a recombination frequency of 0.1. Anyway, in this case the two other markers defining the same chromosomal region gave negative lod scores at the same recombination frequency. In most of the cases the affected members of each of the six families shared different alleles showing the absence of cosegregation within each family. An example of the results in the two most representative families is given in Fig 1.
Example of allele segregation for the markers under study in two representative CDA II families (CDAGE and CDACE): the solid squares and circles indicate the affected individuals. The microsatellite markers are tightly linked to the following candidate genes: α-Man II, mapping on chromosome 5 (5q2.1-2.2); α-Man IIx isozyme, mapping on chromosome 15q25; and human GnT II, mapping on chromosome 14q21. The above-mentioned markers are listed according to the order established in Weissenbach et al21 and Gyapay et al.22
Example of allele segregation for the markers under study in two representative CDA II families (CDAGE and CDACE): the solid squares and circles indicate the affected individuals. The microsatellite markers are tightly linked to the following candidate genes: α-Man II, mapping on chromosome 5 (5q2.1-2.2); α-Man IIx isozyme, mapping on chromosome 15q25; and human GnT II, mapping on chromosome 14q21. The above-mentioned markers are listed according to the order established in Weissenbach et al21 and Gyapay et al.22
Negative results obtained indicated that most likely none of the investigated regions contains the gene involved in CDA-II.
DISCUSSION
CDA-II is a rare recessive disorder often characterized by a mild clinical phenotype. Clinical findings usually do not appear in the neonatal period. For this reason, the possible existence of tissue-specific or developmental stage-specific isozymes should be considered.13,24 A number of associations with other RBC-inherited disorders have been reported and it appears to be clear that the association could worsen clinical findings.25 26 The latter could account for the clinical heterogeneity of this disease.
The Ham's test positivity, the increased agglutinability anti-i, and the SDS-PAGE aspect of band 3 strongly suggest the existence of a glysosylation defect. The formation of such unusual oligosaccharides corresponds to reduced activity of GnT-II14 in some cases and with low α-Man II in others.15 Fukuda et al17 described a variant of CDA-II characterized by the absence of polylactosamines either on protein or on lipids. In this case, a quantitative defect of galactosyltransferase was established.
The glycosylation defect causes hypoglycosylation of band 3; the latter, in turn, causes abnormal clustering of these molecules only during the intracytoplasmic life of this molecule,27 whereas the effect on the anion transport is not well established.28,29 Mapping and cloning of cDNA encoding for GnT-II and α-Man II allow for the molecular studies.18-20 Low expression of α-Man II was shown by Northern blotting and no abnormal mRNA species are detected.15 Sequencing of the coding sequence suggested that a mutation in the promoter region of α-Man II gene results in insufficient transcription.15 Using a previously isolated murine cDNA clone as a probe, Misago et al19 isolated cDNA clones encompassing the human α-Man II cDNA open reading frame and initiated isolation of human genomic clones. This gene maps at 5q21-22 region and encodes a protein of 1144 amino acid residues. A related gene was recently isolated that encodes a truncated protein with 796 amino acid residues and maps at 15q25. The human GnT II is a single exon gene that contains a 1341-bp open reading frame encoding a 447 amino acid protein. This gene was mapped by fluorescence in situ hybridization to chromosome band 14q21.20
Localization of the putative genes and the availability of polymorphic marker loci in these regions allow a linkage study in families with multiple affected siblings.
Our data on linkage analysis and allele segregation showed that there was no linkage between CDA-II phenotypes and the chromosomal regions containing the three candidate genes. These results showed for the first time that the molecular lesion(s) causing CDA-II is not primarily located in α-Man II and GnT II genes.
However, biochemical assays showed a lowered activity of these two enzymatic activities in CDA-II RBCs. In addition, Northern analysis showed in some of our kindreds a reduction of α-Man II and GnT II mRNAs in the same patients, whereas this finding was not detected in the corresponding obligate carriers (A. D'Agostaro, personal communication, September, 1995 and Iolascon et al4 ). These data clearly exclude these genes as candidates and suggest at the same time that the disease is most likely due to a defect of a transcriptional factor regulating both α-Man II and GnT II.
A genome-wide search by means of a large number of CDA II families could allow for mapping of this disease gene. This knowledge will help significantly in our understanding of its pathogenesis and will be essential in the design of a rational therapy.
ACKNOWLEDGMENT
We are grateful to the Associazione Italiana per la Lotta al Neuroblastoma.
Supported by partial funding from the Italian Ministry of Health and by Associazione Italiana per la Ricerca sul Cancro (AIRC), MURST, and Consiglio Nazionale delle ricerche (PF. ACRO, Italy).
Address reprint requests to Achille Iolascon, MD, PhD, Dipartimento di Biomedicina dell'Età Evolutiva, Piazza G. Cesare 11, 70124 Bari, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal