Abstract
Recombinant adeno-associated viruses (rAAV) have been proposed to be gene transfer vehicles for hematopoietic stem cells with advantages over other virus-based systems due to their high titers and relative lack of dependence on cell cycle for target cell integration. We evaluated rAAV vector containing a LacZ reporter gene under the control of a cytomegalovirus (CMV) promoter in the context of primary human CD34+CD2− progenitor cells induced to undergo T-cell differentiation using an in vitro T-lymphopoiesis system. Target cells from either adult bone marrow or umbilical cord blood were efficiently transduced, and 71% to 79% CD2+ cells expressed a LacZ marker gene mRNA and produced LacZ-encoded protein after exposure to rAAV-CMV-LacZ. The impact of transgene expression on the differentiation of T cells was assessed by sequential quantitation of immunophenotypic subsets of virus-exposed cells and no alteration was noted compared with control. The durability of transgene expression was assessed and found to decay by day 35 with kinetics dependent on the multiplicity of infection. In addition, vector DNA was absent from CD4 or CD8 subselected CD3+ cells by DNA-polymerase chain reaction. These data suggest that rAAV vectors may result in robust transgene expression in primitive cells undergoing T-cell lineage commitment without toxicity or alteration in the pattern of T-cell differentiation. However, expression is transient and integration of the transgene unlikely. Recombinant AAV vectors are potentially valuable gene transfer tools for the genetic manipulation of events during T-cell ontogony but their potential in gene therapy strategies for diseases such as acquired immunodeficiency syndrome is limited.
GENE TRANSFER into primary human hematopoietic stem cells faces several challenges affecting clinical application.1,2 Among these are methods of efficient gene transduction, gene expression posttransduction, and the developmental consequences of gene transduction into primitive cells. We have recently developed a system that induces T-cell differentiation from primitive human bone marrow CD34+CD2− or CD34+CD38−CD2− progenitor cells and provides a model by which to address some of these areas specifically as they relate to T-lineage events.3 Such a system has particular relevance to the goal of providing genetically based defenses against human immunodeficiency virus (HIV) infection.
In this study, we have evaluated the efficiency of transduction, transduced gene expression, and the impact of such gene expression with the T-lymphopoiesis model using recombinant adeno-associated virus (rAAV) vectors as a gene delivery system.4,5 This vector type was chosen because, unlike retroviruses, it does not appear to be restricted to dividing cells and, therefore, may be advantageous for hematopoietic stem cells.6-9 Transduction of primary human and murine bone marrow progenitors by rAAV with persistent reporter gene expression has been reported in short-term cultures and animal models.10-14 Furthermore, AAV is a dependent parvovirus and is completely nonpathogenic in the absence of adenovirus or certain members of the herpes virus family.15 Without coinfection with helper virus, the wild-type AAV genome has been reported to integrate stably via its termini into the host cell genome in a site-specific manner and remains latent.16-20 In addition, AAV offers the further advantage over other viral systems such as recombinant retroviruses in that it has a broad host range for infectivity21 and viral preparations at titers in excess of 109 infectious particles per milliliter can be generated.22 These preparations have been used to transduce enriched human CD34+ cell lines at high efficiency and show regulated transgene expression from integrated virus after 2 weeks of cultivation under selection pressure.7 However, rAAV constructs integrate into the host chromosome in a nonspecific fashion, which is suggestive of an alternative mechanism of integration from wild-type AAV.14 Recent studies evaluating rAAV transduction in the absence of selection over prolonged time periods have shown transient expression and low integration levels of rAAV in rapidly dividing cell lines and primary CD34+ cells.23,24 Although transfection of rAAV plasmids and transgene expression in primary T lymphocytes and T-cell lines has been demonstrated,25 the ability of rAAV vectors to sustain long-term transgene expression during T lymphopoiesis remains untested.
We have recently shown that human CD34+CD2− progenitor cells can be induced to undergo T lymphopoiesis on thymic stroma monolayers supplemented with cytokines.3 During the process of T-cell ontogony in vitro, developing cells undergo a series of immunophenotypic changes that are discrete and measurable by fluorescent-activated antibody staining techniques. Progenitor cells transition through a defined intermediate stage in which surface expression of both CD4 and CD8 can be detected coincident with surface CD3 before maturing to CD3+ cells that express either CD4 or CD8 alone. Fluorescence-activated cell sorting (FACS) analysis of sampled cells showed that primitive hematopoietic cells acquire an intermediate phenotype after 2 to 3 weeks of cultivation and require a further 2 to 3 weeks to develop a mature T-cell immunophenotype. Output cells express nuclear RAG2, a polyclonal TCR-Vβ repertoire, which can be infected by HIV-IIIB and have immunofunction as assessed by interleukin-2 (IL-2) production and alloreactivity assays (Freedman et al,3 Rosenzweig et al,26 and J.P.G, unpublished data).
The objectives of this study were to evaluate the effects of rAAV-based gene transduction on the T cells differentiated from primary human progenitors in the culture system and to assess the levels of transduction and expression of a reporter gene, LacZ, by cellular and molecular techniques. This study provides a novel assessment of the temporal expression of an rAAV transgene in primary human hematopoietic progenitor cells differentiating along the T-cell lineage. The data show that gene transduction and expression after transduction with rAAV occurs at high efficiency for developing T cells in short-term cultures and does not perturb the differentiation program, but expression is dramatically reduced at later time points. Although these findings may preclude the use of rAAV vectors as a delivery system for therapy in which long-term expression is necessary, this system can be used to assess the impact of transfer of novel genes on the early stages of T-cell development.
MATERIALS AND METHODS
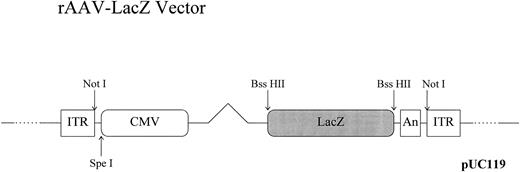
Plasmids and vectors. The rAAV-cytomegalovirus (CMV)-LacZ vector was constructed by placing the Escherichia coli LacZ gene, encoding β-galactosidase, under the transcriptional control of the CMV immediate early promoter as previously described (Fig 1).27 Subconfluent 293 cells were cultured in complete Dulbecco's modified Eagle's medium (BioWhittaker, Walkersville, MD) containing 4.5 g/L glucose, 10% heat-inactivated fetal calf serum (Hyclone, Logan, UT), 2 mmol/L glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin and cotransfected by calcium phosphate precipitation28 with two plasmids. The first encoded both the AAV-LacZ vector and the AAV helper sequences, and the second encoded adenoviral functions necessary for AAV vector production. The adenoviral helper plasmid carries the E2A, E4, and VA RNA regions from adenovirus type 5 and, when transfected, is functionally equivalent to an adenovirus infection with respect to AAV production.29 The transfection was performed according to the method of Wigler et al,28 and AAV vector production was performed in the absence of adenovirus. AAV-LacZ vector purification was performed by cesium chloride isopyknic gradient centrifugation as previously described,27 except that the final adenovirus heat-inactivation step was omitted. The purified, DNase-treated AAV-LacZ vector preparation was titered by X-gal staining of infected 293 cell cultures and by quantitative dot blot assays as described before27 and was routinely in the range of 1012 to 1013 particles/mL. In our experience, the particle-to-transduction unit ratio was between 102 and 103, and subsequent multiplicities of infections (MOI) for progenitor cell experiments were calculated using effective transduction units.
Recombinant AAV-CMV-LacZ vector. LacZ expression cassette was inserted between the ITRs (inverted terminal repeats) of a pUC-based plasmid containing both the AAV ITRs and the AAV rep and cap genes outside the ITRs. Cloning sites used for vector construction are shown. CMV, CMV promoter; An, simian virus 40 (SV40) early polyadenylation signal.
Recombinant AAV-CMV-LacZ vector. LacZ expression cassette was inserted between the ITRs (inverted terminal repeats) of a pUC-based plasmid containing both the AAV ITRs and the AAV rep and cap genes outside the ITRs. Cloning sites used for vector construction are shown. CMV, CMV promoter; An, simian virus 40 (SV40) early polyadenylation signal.
Preparation of thymic stroma cultures. Thymus glands were removed from electively aborted fetuses of 18 to 22 weeks of gestation after informed maternal consent was obtained. Tissue was carefully disaggregated, passed through a mesh sieve, and washed twice in phosphate-buffered saline (PBS). Typical yields were 1 to 10 × 108 cells per gland at greater than 90% viability as assessed by trypan blue exclusion. Cells were resuspended in complete medium: Iscove's modified Dulbecco's medium (Mediatech, Washington, DC) containing 20% fetal calf serum (Sigma, St Louis, MO), glutamine (1 mmol/L), penicillin (10 IU/mL), and streptomycin (10 mg/mL). Aliquots were dispensed into 24-well tissue culture plates at a density of 2 × 106 per well and were incubated at 37°C/5% CO2 until confluent, followed by irradiation (15 Gy). The combination of vigorous washing and irradiation was sufficient to ensure the complete removal of all fetal thymocytes from stroma layers, as previously reported.3
Purification of progenitor cells. Bone marrow samples were obtained in sodium heparin with informed consent by institutional review board (IRB)-approved protocols from healthy adult volunteers by iliac crest aspiration. Umbilical cord blood was obtained according to IRB-approved guidelines. Specimens were collected in sterile bags containing citrate phosphate dextrose anticoagulant (Sigma), transported at room temperature, and used within 48 hours. Mononuclear cells (MNCs) were separated by density gradient centrifugation over Ficoll-Paque (Pharmacia Biotech Inc, Piscataway, NJ), washed twice, and remaining erythrocytes were removed by lysis with 0.15 mol/L ammonium chloride. Cells were resuspended in complete medium and cultured overnight at 37°C to remove adherent cells. The CD34+CD2− MNC fraction was isolated by either sequential immunomagnetic bead selection using anti-CD34 DETACHaBEADs (Dynal, Lake Success, NY) followed by depletion with CD2 beads or sorted using a FACS Vantage cell sorter (Becton Dickinson, San Jose, CA) after staining with anti-CD34 (Amac Inc, Westbrook, ME) and anti-CD2 (Exalpha Corp, Boston, MA) monoclonal antibodies.
T-lymphopoiesis assay. T cells were generated from progenitors as described,3 but with minor modifications. Briefly, 1 to 5 × 104 CD34+CD2− cells per well added to washed, irradiated thymic stromal feeder layers and cultured for a further 3 weeks at 37°C/5% CO2 in complete medium supplemented with IL-12 (R&D Systems, Minneapolis, MN) at 10 ng/mL and flk-2/flt-3 ligand (gift of Dr Stuart Lyman, Immunex, Seattle, WA) at 100 ng/mL. To stimulate expansion of T cells after 3 to 4 weeks, cells were harvested from the thymic stromal monolayers and cultured in complete medium supplemented with IL-2 (Schiapparelli Biosystems, Columbia, MD) at 30 IU/mL and phytohemagglutinin (PHA; Murex, Dartford, UK) at 0.25 μg/mL for a further 2 to 3 weeks. Cultures were replenished twice weekly with cytokine-supplemented complete medium.
Transduction. Recombinant AAV containing a LacZ reporter gene driven by a modified CMV promoter (Fig 1) was prepared by freeze-thaw lysis and used immediately. CD34+CD2− cells (1 to 5 × 104/mL) were exposed to AAV-LacZ supernatant in 1 mL final volume at MOIs ranging from 1 to 10 using vector volumes from 0.5 to 5.0 μL, depending on the vector titer and particular experiment. Transductions were routinely performed on irradiated thymus monolayers immediately after CD34+CD2− isolation by cocultivation with rAAV in complete medium with IL-12 (30 IU/mL) and flk-2/flt-3 ligand (100 ng/mL). Preliminary findings showed that 2 hours of exposure of cells to rAAV was sufficient for maximal transduction and was independent of inclusion of thymus or virus-removal procedures. To simplify the protocol, cells were cocultured directly on the stroma monolayers with AAV, which was washed out by subsequent medium changes, and only cytokines were replenished as described above. Control cells were mock-transduced with an equivolume of culture medium and cytokines alone.
Immunophenotyping of cultured cells by flow cytometry. Cells were harvested by gentle aspiration, washed twice in PBS, and counted, and 2 to 5 × 105 cells were directly stained in a final volume of 100 μL with the following antibodies in the presence of 2% mouse serum (Dako, Carpentiera, CA): fluorescein isothiocyanate (FITC)-anti-CD34 (Amac), FITC-anti-CD2, anti-CD3, anti-CD8; phycoerythrin (PE)-anti-CD2, anti-CD3, anti-CD4, anti-CD8 (Exalpha, Boston, MA). FITC- and PE-conjugated mouse isotype control antibodies were used for each culture. Stained samples were washed three times in PBS and flow cytometric analysis was performed using a FACScan cytometer (Becton Dickinson) immediately or after fixation with fresh paraformaldehyde (2%).
Cytochemical staining for β-galactosidase activity. Duplicate samples of cells from the T-lymphopoiesis assay were harvested by gentle aspiration and washed twice in PBS. Each sample was transferred into three wells of a 96-well plate at 5 × 103 cells per well in complete medium with fresh 5-bromo-4-chloro-3-indoyl β-D-galactopyranoside (X-gal; Sigma) at 150 μg/mL. Plates were incubated overnight at 37°C/5% CO2 in a dark, humidified incubator before examination by phase contrast microscopy. The number of blue cells staining positively with X-gal was enumerated by triplicate counts of at least 500 cells in total from each well, and the percentage of cells expressing β-galactosidase was computed. The mean value and standard error of the mean was calculated for comparative analyses, and a two-tailed Student's t-test was used to determine statistical significance.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis. mRNA was prepared from between 103 and 106 sorted or cultured cells using guanidium thiocyanate and oligo dT spun columns (QuickPrep mRNA purification kit; Pharmacia) before DNaseI (Promega, Madison, WI) digestion. Random hexanucleotide primers and Moloney reverse transcriptase (GIBCO-BRL, Grand Island, NY) were used to prepare cDNA, which was stored at −20°C. Amplification was performed using one quarter of the cDNA product in each PCR reaction (50 μL) with 2.5 U Taq DNA polymerase (Pharmacia) and 0.4 μmol/L of each of the following oligonucleotide sequences: LacZ: 3′ primer, GAC ACC AGA CCA ACT GGT AAT G; 5′ primer, CTG AAT ATC GAC GGT TTC C; controls were as follows: β-actin: 3′ primer, GTG GGG CGC CCC AGG CAC CA; 5′ primer, GTC CTT AAT GTC ACG CAC GAT TCC; and Gsα (a constitutively expressed G protein30 ): 3′ primer, GCT GCT GGC CAC CAC GAA GAT GAT; 5′ primer, GTG ATC AAG CAG GCT GAC TAT GTG.
PCR amplification was performed in a Gene Amp 9600 thermal cycler (Perkin Elmer Corp, Norwalk, CT) with an initial denaturation (95°C for 5 minutes) followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 30 seconds, and extension at 72°C for 60 seconds for the LacZ and β-actin amplifications; 30 cycles of 94°C, 55°C, and 72°C for 1 minute each were used for the Gsα amplification. A single 10-minute extension after amplification was performed at 72°C for all reactions. PCR products (10 μL) were electrophoretically separated on ethidium bromide-stained agarose (1.5%) gels and photographed under UV light. Specificity was confirmed by Southern blot hybridization at 55°C for 2 hours using Expresshyb hybridization solution (Clontech, Palo Alto, CA) with probes labeled with digoxigenin or 32P. Sequences of internal probes were CTG AAC CCT AAG GCC AAC CGT G for LacZ and TCA GTA TCG GCG GAA TTC CAG CTG A for β-actin.
Blots were washed under high stringency, followed by incubation with an antidigoxigenin antibody alkaline phophatase conjugate and detection by chemiluminescence or autoradiography as appropriate. Parallel extractions were performed using a HeLa cell line stably transfected with LTR-LacZ gene31 as a positive control and samples without RT or water alone were used to exclude DNA contamination.
DNA analysis. High molecular weight DNA was isolated from washed bulk and sorted cells by sequential cell lysis, RnaseA treatment, and protein precipitation according to the Puregene kit instructions (Gentra, Minneapolis, MN). Hydrated DNA from equivalent cell numbers was subjected to PCR analysis in parallel to RT-PCR. DNA was extracted from a HeLa cell line expressing LacZ as a positive control,31 and water alone was used as a negative control for all subsequent PCRs.
RESULTS
In vitro T-cell differentiation is not perturbed by rAAV transduction. For gene transduction of primitive cells to become a feasible methodology for stem cell gene therapy, gene transfer must occur without perturbing the ability of primitive cells to develop along the T-lymphoid lineage. Therefore, the aim of initial studies was to assess if the pattern of T-cell differentiation in our system, which closely recapitulates intrathymic T-cell ontogeny in vivo, was perturbed by rAAV transduction. The input cells for the T-lymphopoiesis assay were isolated from adult bone marrow or umbilical cord blood MNCs using either fluorescence-activated cell sorting or immunomagnetic beads (Fig 2A). Flow cytometric analysis was performed by gating forward and side scatter signals typical of hematopoietic progenitor cells and contained greater than 90% CD34+ cells and less than 2% residual CD2+ immature T-cell precursors. The highly enriched progenitor cell fraction was cultured on thymic stroma with rAAV containing a CMV-LacZ transgene at an MOI of 10 or mock-transduced with medium alone as a control.
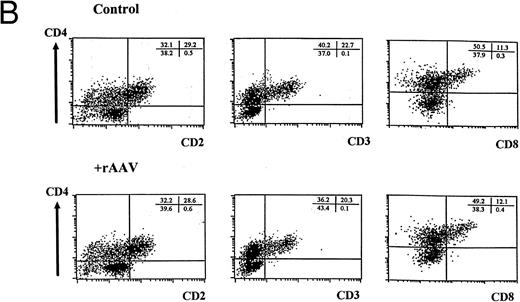
FACS analysis of cells differentiating in the T-lymphopoiesis system. (A) Input cells: top panel is forward scatter (Fsc) versus side scatter (Ssc) profile showing the gate (G1) used for analysis of lymphocyte-like cells. The middle panel is a flow cytometric profile of surface CD2 and CD34 expression of MNCs used for flow cytometric purification. The R1 gate (1% to 2% MNCs) was established to sort CD34+CD2− cells by comparing positive and negative stained controls. The lower panel is a representative flow cytometric analysis of CD34+CD2− cells isolated by flow cytometry or magnetic bead separation before culture on thymic stroma. Quadrants were set using matched isotype controls, and comparable purification results were obtained in multiple independent experiments. (B) Differentiation of cells expressing surface CD2, CD3, CD4, and CD8 in human thymic stroma culture is not perturbed by exposure to rAAV. FACS analysis of CD34+CD2− cells either mock-transduced (upper panel) or exposed to rAAV (lower panel) after 21 days in the T-lymphopoiesis system. Quadrants were established using matched isotype antibody controls and represent one of three comparisons. Minor cell populations expressing nonlymphoid phenotypes were detectable at early time points, but these were absent at later time points (data not shown).
FACS analysis of cells differentiating in the T-lymphopoiesis system. (A) Input cells: top panel is forward scatter (Fsc) versus side scatter (Ssc) profile showing the gate (G1) used for analysis of lymphocyte-like cells. The middle panel is a flow cytometric profile of surface CD2 and CD34 expression of MNCs used for flow cytometric purification. The R1 gate (1% to 2% MNCs) was established to sort CD34+CD2− cells by comparing positive and negative stained controls. The lower panel is a representative flow cytometric analysis of CD34+CD2− cells isolated by flow cytometry or magnetic bead separation before culture on thymic stroma. Quadrants were set using matched isotype controls, and comparable purification results were obtained in multiple independent experiments. (B) Differentiation of cells expressing surface CD2, CD3, CD4, and CD8 in human thymic stroma culture is not perturbed by exposure to rAAV. FACS analysis of CD34+CD2− cells either mock-transduced (upper panel) or exposed to rAAV (lower panel) after 21 days in the T-lymphopoiesis system. Quadrants were established using matched isotype antibody controls and represent one of three comparisons. Minor cell populations expressing nonlymphoid phenotypes were detectable at early time points, but these were absent at later time points (data not shown).
After 21 days of culture, we observed a 10- to 20-fold increase in cell number in parallel with the appearance of a population of intermediate CD3+ T cells expressing both CD4 and CD8 (Fig 2B). Cultivation with rAAV did not affect cellular expansion as assessed by duplicate cell counts on replicate wells (Table 1). Acquisition of T-cell immunophenotype was similarly not perturbed, and the proportion of cells expressing T-cell markers was indistinguishable from controls (Fig 2B and Table 1). No differences were observed when these parameters were evaluated at different time points in independent experiments (data not shown). In addition, RT-PCR of developing T cells showed that RAG2 and TCRαβ expression was indistinguishable between control and rAAV-transduced cultures (data not shown).
Effect of rAAV Infection on Proliferation and Immunophenotype of Cells Developing in the T-Lymphopoiesis System
| . | . | Expansion Index . | Immunophenotype . | |||
|---|---|---|---|---|---|---|
| . | . | . | CD2+ . | CD3+ . | CD4+ . | CD8+ . |
| Day 21 | Control | 11.6 ± 4.4 | 29.2 ± 2.0 | 18.9 ± 5.5 | 51.8 ± 12.7 | 9.8 ± 2.5 |
| +rAAV | 12.4 ± 3.6 | 24.7 ± 5.4 | 16.7 ± 5.2 | 50.7 ± 11.2 | 10.8 ± 2.5 | |
| Day 35 | Control | 86.3 ± 65.4 | 73.5 ± 5.8 | 89.1 ± 4.7 | 87.4 ± 4.3 | 10.5 ± 3.7 |
| +rAAV | 82.0 ± 53.7 | 68.7 ± 4.3 | 85.4 ± 6.1 | 83.9 ± 3.9 | 14.0 ± 5.3 | |
| . | . | Expansion Index . | Immunophenotype . | |||
|---|---|---|---|---|---|---|
| . | . | . | CD2+ . | CD3+ . | CD4+ . | CD8+ . |
| Day 21 | Control | 11.6 ± 4.4 | 29.2 ± 2.0 | 18.9 ± 5.5 | 51.8 ± 12.7 | 9.8 ± 2.5 |
| +rAAV | 12.4 ± 3.6 | 24.7 ± 5.4 | 16.7 ± 5.2 | 50.7 ± 11.2 | 10.8 ± 2.5 | |
| Day 35 | Control | 86.3 ± 65.4 | 73.5 ± 5.8 | 89.1 ± 4.7 | 87.4 ± 4.3 | 10.5 ± 3.7 |
| +rAAV | 82.0 ± 53.7 | 68.7 ± 4.3 | 85.4 ± 6.1 | 83.9 ± 3.9 | 14.0 ± 5.3 | |
The expansion index was determined by dividing the number of viable cells present at different time points by the number of starting CD34+CD2− cells from independent experiments (n = 6). Trypan blue staining was used to evaluate cell viability, which was routinely greater than 90%. Immunophenotype was evaluated for a subset of the experiments (n = 3) by fluorescent antibody staining and FACS-quadrant analysis as described for Fig 2. Total percentages of cells expressing individual antigens are shown, and the dominant cell population present at day 21 was CD2+CD3+CD4+, whereas at day 35, maturation to single-positive CD3+CD4+ and CD3+CD8+ T cells had occurred. All data are the mean ± SD from duplicate readings.
These data indicate that exposure to rAAV was not toxic to T cells developing from hematopoietic progenitors and that the differentiation program of T cells was not intrinsically altered by the transduction process.
Transfer and expression of a reporter gene in differentiating T cells. Genes inserted into the primitive pluripotent cells must be transcriptionally active during T-cell differentiation for stem cell gene therapy to protect progenitor cells from HIV infection. We used the T-lymphopoiesis assay to examine expression of a reporter gene, LacZ, as progenitor cells acquired T-cell immunophenotypic characteristics. Enriched CD34+ CD2− cells were transduced with rAAV or mock-transduced with medium alone as control and cultured for 21 days on thymic stroma. Immature CD2+ T cells were sorted by fluorescence-activated cytometry at greater than 90% purity (data not shown) and tested for LacZ expression at the cellular level by X-gal staining or at the transcriptional level by RT-PCR.
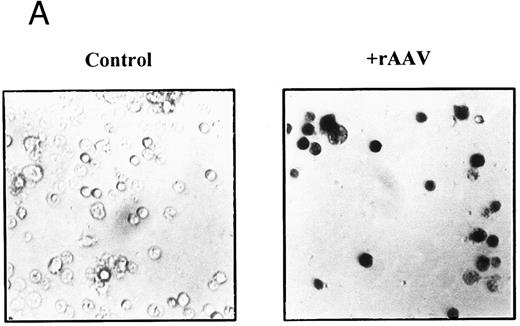
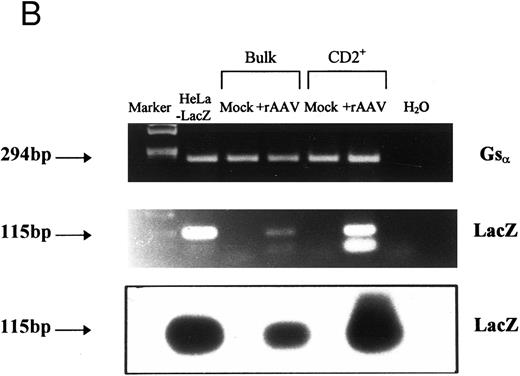
In three independent experiments, we found that 74.7% ± 2.3% of cells in the CD2+ fraction stained positively with X-gal after 21 days, but there was no β-galactosidase activity in mock-transduced controls (Fig 3A). This pattern of staining correlated with mRNA expression when we tested aliquots of the CD2+ sorted cells by RT-PCR (Fig 3B). A 115-bp PCR product was amplified by LacZ-specific primers from cDNA isolated from rAAV-transduced CD2+ cells at a level comparable to a cell line stably transfected with LacZ gene. The identity of the product was confirmed by Southern blot hybridization with a LacZ-specific probe (Fig 3B), and this was a consistent finding for all experiments at early time points. Nontransduced cells and controls without RT were negative for LacZ expression, and all cellular samples were positive for constitutively expressed genes, β-actin, or Gsα . Gsα PCR products permit discrimination of cDNA from genomic DNA by the size of the PCR products; only samples positive for Gsα cDNA and negative for genomic DNA were included in the analysis.
Expression of LacZ transgene in developing T cells after rAAV-mediated gene transfer. CD34+CD2− cells were cultured on thymic stroma either with rAAV-LacZ at MOI of 10 or medium alone (control). CD2+ cells were FACS purified after 21 days and aliquotted for analysis of expression. (A) β-galactosidase activity: triplicate samples of 5 × 103 cells per well were incubated with X-gal at 150 μg/mL and enumerated by phase contrast microscopy with ×40 objective magnification. (B) Detection of LacZ mRNA in bulk or CD2+ T cells by RT-PCR. mRNA was prepared from equivalent numbers of cells and cDNA generated by RT with random hexamers. The products were amplified by PCR with primers specific for an internal region of LacZ or Gsα . Specificity of the PCR products was confirmed by Southern blot hybridization with a 32P-labeled probe specific for LacZ. A Hela cell line stably transfected with LTR-LacZ was used as a positive control, and analysis was repeated for three independent experiments.
Expression of LacZ transgene in developing T cells after rAAV-mediated gene transfer. CD34+CD2− cells were cultured on thymic stroma either with rAAV-LacZ at MOI of 10 or medium alone (control). CD2+ cells were FACS purified after 21 days and aliquotted for analysis of expression. (A) β-galactosidase activity: triplicate samples of 5 × 103 cells per well were incubated with X-gal at 150 μg/mL and enumerated by phase contrast microscopy with ×40 objective magnification. (B) Detection of LacZ mRNA in bulk or CD2+ T cells by RT-PCR. mRNA was prepared from equivalent numbers of cells and cDNA generated by RT with random hexamers. The products were amplified by PCR with primers specific for an internal region of LacZ or Gsα . Specificity of the PCR products was confirmed by Southern blot hybridization with a 32P-labeled probe specific for LacZ. A Hela cell line stably transfected with LTR-LacZ was used as a positive control, and analysis was repeated for three independent experiments.
These data show a high fraction of cells expressing LacZ, suggesting either high efficiency gene transfer or preferential outgrowth of transduced cells. Encouraged by the large population of cells expressing the rAAV transgene in developing T cells, we assayed the long-term expression of LacZ in cells sequentially isolated from the T-cell system.
Expression of rAAV-LacZ is reduced at later time points in T lymphopoiesis. For rAAV to be an efficient delivery vehicle for stem cell gene therapy, introduced genes must exhibit persistent expression in mature T cells. Recombinant AAV can exist in two forms in the cell that are both permissive for expression. The DNA-integrated form within the cell nucleus is associated with long-term expression in other tissue systems,17,32 and the episomal, cytoplasmic form is associated with short-term expression only, because it will segregate equivalently into daughter progeny at cell division and be lost over time.24
We decided to use two complementary, yet independent approaches to address these questions in our system. In the cell biologic approach, we predicted that, if the rAAV transgene was integrated into host cell DNA, expression of LacZ and the presence of β-galactosidase would be maintained at later time points and this effect would be independent of MOI. However, if developing T cells were transduced with rAAV-LacZ in an episomal form, then we hypothesized that levels of expressed LacZ and β-galactosidase would diminish after the intense cell proliferation in the system and this would occur more rapidly at lower MOIs.
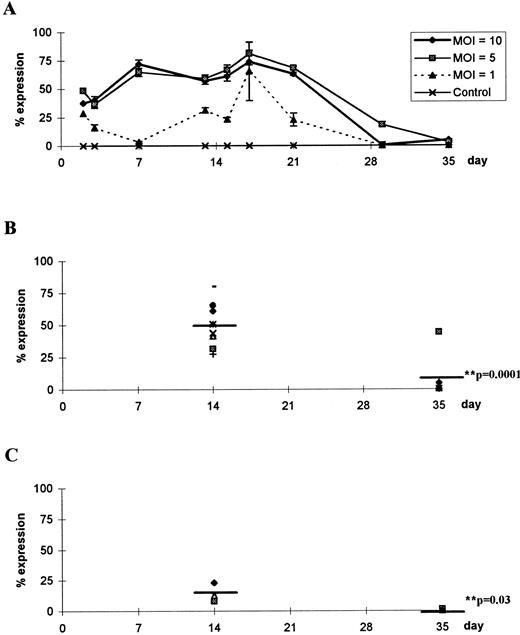
To test this hypothesis, we assayed cells for β-galactosidase activity at various time points from 3 days to 5 weeks of cultivation of CD34+CD2− cells on thymic stroma (Fig 4A). X-gal staining showed that β-galactosidase levels were sustained at high efficiency up to 3 weeks and then declined to undetectable levels when examined at 5 weeks. This was a consistent finding for 7 of 8 independent experiments in which X-gal staining at early time points showed that between 50% and 80% of cells were positive for β-galactosidase and decreased rapidly to 0% after 5 weeks (Fig 4B). The reduction at later time points was statistically significant compared with early expression when assessed with an unpaired two-tailed t-test (P = .0001; Fig 4B). Although overall transduction efficiency was reduced at lower MOI, the decline in expression at later time points was consistent and statistically significant (P = .03; Fig 4C).
Time course of β-galactosidase activity during T lymphopoiesis. (A) CD34+CD2− cells were transduced at MOIs of 10, 5, and 1 or were mock-transduced with an equivolume of medium (control). Two samples were taken at each time point and independently incubated with X-gal at 150 μg/mL, and the number of cells staining positively was enumerated. Data are mean values ± SEM of triplicate readings in a representative experiment. Data from seven independent experiments were analyzed after 14 days or 35 days to compare expression of β-galactosidase after infection with rAAV at (B) MOI = 10 or (C) MOI = 1. Mean values are denoted by a straight line and P values were calculated using a two-tailed unpaired t-test.
Time course of β-galactosidase activity during T lymphopoiesis. (A) CD34+CD2− cells were transduced at MOIs of 10, 5, and 1 or were mock-transduced with an equivolume of medium (control). Two samples were taken at each time point and independently incubated with X-gal at 150 μg/mL, and the number of cells staining positively was enumerated. Data are mean values ± SEM of triplicate readings in a representative experiment. Data from seven independent experiments were analyzed after 14 days or 35 days to compare expression of β-galactosidase after infection with rAAV at (B) MOI = 10 or (C) MOI = 1. Mean values are denoted by a straight line and P values were calculated using a two-tailed unpaired t-test.
We reasoned that reduction of LacZ expression may be attributable to AAV-induced toxicity at later time points and selective loss of transduced cells. However, using trypan blue staining the isolated cells and parallel FACS analysis, we found no difference in cell viability or immunophenotypes between control and transduced (Table 1).
Taken together, these findings suggested that rAAV may exist in an episomal state without integration in developing T cells. To establish the molecular basis of this conclusion, we examined the cellular DNA of cells sampled from the T-lymphopoiesis system by PCR and Southern blot analysis.
Molecular characterization of the rAAV vector in developing T cells. The observations that developing T cells could be transduced efficiently and that expression persisted for up to 21 days before rapidly declining could have at least three explanations. First, rAAV-LacZ was integrated into host DNA, but was not transcriptionally expressed at later time points. Second, integration occurred, but expression was ablated at a posttranscriptional level. Third, rAAV was not integrated but transiently existed in the episomal form.
To distinguish between these possibilities, we analyzed the relationship of the rAAV genome relative to the host chromosomal DNA at various time points, taking advantage of the expected size differential of integrated versus episomal viral DNA. High molecular weight DNA was isolated from cells in independent experiments at an intermediate (2 weeks) and later time point (5 weeks). In addition, aliquots of cells were used to assess transgene mRNA presence by RT-PCR and protein expression and activity by X-gal staining. The DNA samples were amplified by PCR using LacZ-specific primers and characterized by Southern analysis using an internal LacZ-specific probe, recognizing that trapping of smaller DNA species may occur in the high molecular weight DNA preparation leading to false-positive results when analyzed by PCR. Therefore, positive results from this type of assay may not be definitive due to amplification from concatameric rAAV, whereas negative results would be more reliable. However, Southern blot techniques to characterize viral integration definitively are precluded by the limited numbers of cells that can be attained in this primary cell system, and loss of detection is an indirect measure of depletion of intracellular rAAV in a heterogeneous cell population.
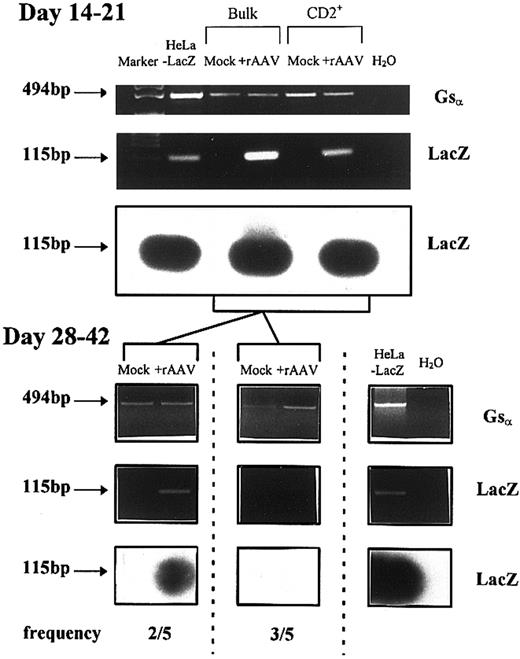
Figure 5 shows that the LacZ transgene sequence was present in the high molecular weight DNA fraction at early time points in all experiments. RT-PCR with LacZ primers and X-gal staining was consistently positive (data not shown). When cells from the same experiments were sampled at later time points (beyond 4 weeks), DNA-PCR remained positive for LacZ in 2 of 5 experiments, but was negative in 3 of 5 experiments (Fig 5). All mRNA samples were positive for constitutively expressed control genes, β-actin, or Gsα , and DNA samples were positive for the large molecular weight PCR product of Gsα .
Analysis of rAAV-LacZ genome in high molecular weight DNA from developing T cells derived from CD34+CD2− isolated at various time points after culture on thymic stroma. Shown are representative data from five experiments at either early time points (14 to 21 days) or later time points (28 to 42 days). PCR was performed with LacZ-specific primers and the identity of the products was confirmed by Southern blotting with a digoxygenin-labeled probe specific to an internal region of LacZ. Gsα primers were used as a positive control for genomic DNA quality and a single 494-bp product was detected.
Analysis of rAAV-LacZ genome in high molecular weight DNA from developing T cells derived from CD34+CD2− isolated at various time points after culture on thymic stroma. Shown are representative data from five experiments at either early time points (14 to 21 days) or later time points (28 to 42 days). PCR was performed with LacZ-specific primers and the identity of the products was confirmed by Southern blotting with a digoxygenin-labeled probe specific to an internal region of LacZ. Gsα primers were used as a positive control for genomic DNA quality and a single 494-bp product was detected.
In the experiments in which DNA-PCR was negative for LacZ, RT-PCR and X-gal staining were both consistently negative, which indicated that rAAV genes were absent and not expressed at later time points. However, when DNA-PCR was positive for LacZ presence, RT-PCR and X-gal staining could be either both positive (in 1 experiment) or both negative (in 1 experiment), implying that transgene sequences were expressed from integrated DNA (the former) or that there was either trapped episomal DNA yielding a false-positive or a dampened expression from an integrated viral genome.
The interexperimental variability observed can be explained if there are differential rates of progenitor cell development into mature T cells in our system. Earlier data have shown that rAAV transgene DNA was present in developing T cells after 2 to 3 weeks of cultivation, and these sequences may persist in some experiments due to the presence of immature T cells in cultures that have not fully differentiated. Late-stage analysis (4 to 5 weeks) of cultures was further complicated by the heterogeneous phenotypes of minor populations of non-T cells copresent with the dominant mature T-cell phenotypes. To clarify our findings, we decided to investigate the integration and expression of rAAV-LacZ vectors in populations of purified mature CD3+ cells.
Lack of expression or integration of a reporter gene in the CD3+-enriched mature CD4+ or CD8+ T-cell fractions. Flow cytometrically purified CD3+ T cells were isolated after 5 weeks of cultivation and the level of expression of the transduced gene was assessed. Cells that had been cultured on thymic stroma for 4 weeks had 8.7% ± 4.6% cells that stained positive for β-galactosidase with X-gal; these cells had been analyzed 2 weeks earlier and were 76% ± 4.6% positive and contained detectable LacZ by RT-PCR and DNA-PCR. Cells derived from this culture were stimulated with IL-2 and PHA for 3 days on thymus to expand mature T-cell populations (Fig 6A). Expansion of single-positive CD4+ and CD8+ T cells was observed and harvested cells were sorted into CD3+CD4+ and CD3+CD8+ fractions at greater than 95% purity. The proportion of T-cell phenotypes and cell numbers isolated from transduced and control cultures after expansion were equivalent (Table 1 and Fig 6A), suggesting that AAV was not toxic to developing progenitors. These subpopulations were tested for LacZ expression and the presence of LacZ DNA in high molecular weight DNA preparations. Expression of LacZ transgene was absent from sorted populations as tested by X-gal staining and RT-PCR analysis (data not shown). Figure 6B shows that DNA-PCR using LacZ primers was negative, and this was confirmed by Southern blot hybridization with a LacZ-specific internal probe. Nontransduced control samples and water controls were negative, and all cellular samples were positive for β-actin. These data support the conclusion that rAAV transgene was neither integrated nor expressed in highly purified mature single-positive T-cell fractions.
Characterization of the rAAV-LacZ DNA in FACS-purified mature CD3+ T cells derived from transduced CD34+CD2− cells. (A) Cells cultured for 28 days expressed surface CD3, CD4, and CD8 and were stimulated with IL-2 and PHA as described in the Materials and Methods. CD3+CD4+ and CD3+CD8+ cells were sorted by flow cytometry using the depicted gates after quadrants were established with matched isotype control antibodies. (B) PCR analysis of high molecular weight DNA extracted from purified mature single-positive T cells. PCR and Southern blot analysis for LacZ transgene was performed as described in Fig 5. Primers for β-actin were used as a positive control. Intervening wells between the last two lanes have been spliced out for clarity.
Characterization of the rAAV-LacZ DNA in FACS-purified mature CD3+ T cells derived from transduced CD34+CD2− cells. (A) Cells cultured for 28 days expressed surface CD3, CD4, and CD8 and were stimulated with IL-2 and PHA as described in the Materials and Methods. CD3+CD4+ and CD3+CD8+ cells were sorted by flow cytometry using the depicted gates after quadrants were established with matched isotype control antibodies. (B) PCR analysis of high molecular weight DNA extracted from purified mature single-positive T cells. PCR and Southern blot analysis for LacZ transgene was performed as described in Fig 5. Primers for β-actin were used as a positive control. Intervening wells between the last two lanes have been spliced out for clarity.
DISCUSSION
Hematopoietic stem cells with long-term repopulating ability are considered to reside in a quiescent state maintaining their ability to reconstitute the blood and immune system under specific physiologic or pathologic conditions. The repopulating potential of these cells makes them highly attractive targets for gene-based approaches to multiple disease states. The concept of molecularly shielding stem cells from HIV-1 infection has theoretic potential as an approach to restore immune function in infected individuals.2 The potential for accomplishing this possibility has acquired greater feasibility as multiple constructs capable of inhibiting the HIV-1 life cycle have been developed and tested in cell lines and primary T cells.33-39
Viral-based gene transfer systems have inherent limitations that may be prohibitive in successfully accomplishing stem cell gene therapy. Traditionally used retroviral constructs are potentially limited by their low titers and the requirement for active cycling of target cells. Attempts to overcome these limitations have included the generation of lentivirus-retrovirus chimeras,39 the development of regulated episomally maintained virus vectors,40 and the use of recombinant adeno-associated viruses.5
Transduction of stem cells, sustained expression of transgenes during differentiation, and the impact of transduction and transgene expression on the biology of the differentiation process remain substantial issues to be addressed in moving this approach forward to clinical testing. However, the recent development of in vitro human T-cell differentiation systems from adult bone marrow progenitor cells3 26 has provided the basis to be able to assess several of these fundamental issues ex vivo. Using primary human progenitor CD34+CD2− cells isolated from adult bone marrow and umbilical cord blood, we have analyzed the ability of an rAAV vector to transduce primitive cells developing along the T-cell lineage with a reporter gene.
The data presented here show that an rAAV-delivered transgene, LacZ, driven by a CMV promoter can infect primitive progenitor cells with high efficiency and result in robust, transient expression of the transgene in the absence of selection. We found that expression levels of LacZ were dependent on multiplicity of infection and were maximal after 21 days in culture, when the CD2+CD3+CD4+ and CD2+CD3+CD4+CD8+ phenotypes were predominant. The mechanism for this upregulation may be due to developmental stage-specific regulation of rAAV-transgene expression. rAAV infection of primitive cells and expression of the transgene did not induce cell toxicity as measured by cell numbers and viability nor did it perturb the differentiation program of the cells as they acquired the phenotypic and genotypic characteristics of mature single-positive T cells. No differences were observed when CD34+ progenitors from adult bone marrow or fetal cord blood were tested (data not shown). As such, these vectors represent potentially valuable reagents to assess the impact of specific transgenes such as transcription factors41 or other regulatory genes42 on the events in early T-lineage commitment. In addition, they may provide a useful reagent for screening the capacity of specific anti-HIV constructs to blunt HIV infection of cells representing multiple stages of developing T cells. An issue that may have particular importance is the ultimate application of stem cell gene therapy for immune reconstitution.
However, the longevity of expression of the reporter gene in these studies was short (<35 days) and the decay of detectable transgene DNA at low MOI suggests that rAAV may not be capable of providing the long-term transgene expression necessary for a satisfactory stem cell approach. Although extinction of transgene expression is a possible explanation for our findings, the data are most consistent with failure of the rAAV genome to integrate stably. We showed that the temporal kinetics of LacZ expression in proliferating cells were predicted by a model in which stable integration did not occur and transient expression occurred from an episomal vector. Although purified CD2+ T cells consistently contained rAAV vector DNA sequences in the high molecular weight DNA fraction after 21 days, this observation was not predictive of long-term transgene expression. Furthermore, mature T cells (both pooled and sorted CD3+ subpopulations) that developed in the in vitro T-lymphopoiesis system in the majority of experiments had lost vector DNA sequences and did not express LacZ.
The heterogeneous constitution of cell cultures developing on thymic stroma monolayers complicate definition of the linear relationships of accepted true T-cell precursor subpopulations. Although it is formally possible that nontransduced CD34+CD2− progenitors developed into CD2−CD4+ intermediates before differentiating to CD3+CD4+ single-positive mature T cells43 and that we might be observing selective outgrowth of a nontransduced population, the balance of evidence suggests that this is an unlikely event. First, the uniformity of transduction (50% to 80%) of all populations analyzed (initial CD34+CD2− cells, sorted CD2+ cells, and bulk cultures) between day 0 and day 21 and the observation that CD2− cells expressed comparable LacZ at day 21 (data not shown) reduce the possibility that the nontransduced CD2− intermediates are the only precursors of mature CD3+CD4+ and CD3+CD8+ cells. All mature T cells expressed CD2 in our system (data not shown), and the most likely candidate precursor cells are the subpopulations with the immunophenotypes of characterized human thymocytes (CD2+CD4+, CD3−CD4+, CD3−CD4+CD8+, and CD3dim CD4+CD8+) that have been previously shown by others to possess the capacity to differentiate into mature single-positive T cells concomitant with the expression of CD2. In addition, the kinetics of cell growth and relative preservation of subpopulations argue against the selective expansion of nontransduced cells. The data suggest that extinction of nonintegrated episomal vector sequences is a more likely mechanism.
The lack of expression after extended time periods is unlikely to be attributable to a specific reporter gene, because other studies have observed a similar effect with a truncated rat nerve growth factor receptor gene.24 It is more likely that the mechanism of integration is influenced by critical auxiliary AAV virion proteins, including AAV Rep,44 45 which need to be present in sufficient quantities to mediate efficient integration. Further analysis of this mechanism and an increased understanding of the stability of the vector-chromosome junctions is required for this approach to develop.
Furthermore, the variation in outcome in this study is suggestive of limitations in the biology of rAAV vectors that may restrict long-term stable gene expression without selection. In one case in which vector was present in the DNA after 5 weeks of cultivation, expression of transgene was undetectable by RT-PCR and X-gal staining. This indicates that repression of transgene in T-cell progeny may be significant at later time points and a similar phenomenon has recently been reported in T cells developing in SCID-hu mice from CD34+ cells transduced with retroviral constructs.46
In summary, we have shown the temporal course of a purified rAAV vector transduction of human hematopoietic progenitor cells derived from different tissue sources developing along the T-cell lineage without selection. The data indicate that, although rAAV mediates high gene transfer and expression, vector genome analysis suggests that vector DNA is not stably integrated and transgene expression declines over time. In the absence of transgene expression from an integrated genome, transduction of the highly proliferative stem cell is unlikely to be useful for in vivo gene therapy; however, this system offers considerable potential for assessing events early in T-lineage commitment.
ACKNOWLEDGMENT
The authors gratefully acknowledge the assistance of Dr Robert Fallon and the staff at the St Louis University Hospital (St Louis, MO) for providing cord blood samples and thank our bone marrow volunteers for their donations.
Supported by National Institutes of Health Grants No. HL44851 and HL55718 and by the Richard Saltenstall Charitable Trust. J.P.G. is the recipient of a Nessel Gene Therapy Fellowship.
Address reprint requests to David T. Scadden, MD, AIDS Research Center, Massachusetts General Hospital, Bldg 149, 13th St, Charlestown, MA 02129; email: scadden.david@mgh.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal