Abstract
Tumor-derived DNA has been shown in various cell-free body fluids. In this study, soluble tumor-derived DNA was analyzed in serum and plasma samples of patients with B-cell malignancies. DNA was extracted from tumor cell specimens as well as serum and plasma samples collected from 110 patients with non-Hodgkin's lymphoma and acute B-precursor lymphoblastic leukemia and was subjected to polymerase chain reaction (PCR) analysis for rearranged immunoglobulin heavy chain DNA. In 54% of serum or plasma samples analyzed at different times before and during treatment, clonal DNA from a rearranged immunoglobulin heavy chain locus was detectable. When examined at diagnosis and before any treatment, clonotypic DNA was found in serum or plasma of 86% of the patients. Serum or plasma from patients with systemic or bulky disease was uniformly PCR positive, whereas clonotypic DNA was also recovered from the serum or plasma from the majority of patients with limited disease stages. Degradation of clonal DNA by nucleases in vitro was shown to be one cause of false-negative PCR results. This technical drawback can be relieved by adding a nuclease inhibitor like EDTA, ie, by using plasma instead of serum for PCR analysis. Treatment of patients with cytotoxic drugs was followed by rapid clearance of DNA from the peripheral blood, suggesting that soluble tumor-derived DNA might be associated with viable and proliferating tumor cells. Follow-up studies showed a close correlation of persisting soluble tumor-derived DNA with resistant disease or early relapse. In summary, these data suggest that tumor-derived DNA can be detected in serum or plasma of the majority of patients with B-cell malignancies and that testing of serum or plasma for tumor-associated DNA may be a novel parameter for monitoring response to treatment.
THE ABILITY TO detect small numbers of viable tumor cells is considered a major issue in the management of patients with curable malignancies including acute lymphoblastic leukemia (ALL) and intermediate and high grade non-Hodgkin's lymphoma (NHL). The majority of these patients respond to remission induction treatment, but at least 50% of them will ultimately relapse.1 2 With the availability of more aggressive second-line or late intensification treatment protocols, including high-dose chemotherapy/radiotherapy supported by autologous hematopoietic stem cells, the question arises as to how to identify patients earlier on who fail conventional treatment protocols.
Next to prognostic variables at presentation,3 the quality of response to the initial treatment courses is a possible and intuitively logical prognostic factor. Patients who slowly enter complete remission have been reported to survive free of disease for a shorter period of time.4-6 Slow responders may, thus, benefit from aggressive second-line treatment protocols. This concept has been challenged by a study that did not show improved long-term results in slowly responding patients who entered an autologous transplantation program instead of continuing conventional treatment.7 Marrow transplantation may have been an ineffective salvage treatment for these patients. Another reason might be the difficulty to reliably characterize the quality of response in lymphoma patients by conventional imaging techniques.8 9
Molecular studies have significantly contributed to our ability to characterize complete remissions in patients with leukemia.10-15 Lymphoma cells can be detected by similar polymerase chain reaction (PCR) techniques.16 However, with the possible exception of follicular lymphoma,17-20 the value of PCR studies for predicting treatment results in lymphoma is unclear. One reason may be the unsolved question of, What tissue must be used for follow-up analysis? In contrast to lymphoblastic leukemia, in which bone marrow and peripheral blood are the prime tumor-involved tissues, it is uncertain whether these tissues are the right choices in patients with lymphoma. Clinically occult tumor cells can be detected in the bone marrow and blood of most patients with follicular lymphoma,21 but this may not be true for other lymphoma subtypes.
During pilot studies that we have performed with serum and plasma samples from patients with NHL and ALL, it became clear that monoclonal immunoglobulin heavy chain (IgH) complementarity-determining region III (CDRIII) DNA could be amplified from cell-free blood samples of some patients. The size of the PCR products amplified from these sources was identical to the CDRIII products amplified from DNA extracted from the tumor cells of the same patients.22 These observations suggested that serum or plasma may be a valuable source of tumor-derived DNA and, therefore, spurred a systematic evaluation of this phenomenon to answer the following questions: Can clonotypic DNA be shown in the serum or plasma of all patients with B-lymphocyte malignancies? Is there any clinical value of using serum or plasma as a source of tumor-derived DNA?
MATERIALS AND METHODS
Tumor cell specimens, peripheral blood leukocytes, serum, and plasma samples. The study was performed on 293 serum samples and 192 plasma samples collected from 110 patients with B-NHL (n = 75) or B-precursor ALL (n = 35) treated at the Department of Medicine III, University of Ulm Medical Center, Ulm, Germany. A total of 87 serum samples were obtained from a serum bank of the Department of Virology, University of Ulm, and 398 samples were prospectively collected for this study. Plasma samples from 16 patients with myeloma were also included in pilot studies of DNA extraction from plasma. Serum and plasma samples from 37 sex- and age-matched normal volunteers served as controls. Tumor cells were analyzed in all patients whose serum or plasma turned out to be negative by PCR analysis for rearranged CDRIII to exclude uninformative rearrangements. Tumor cell DNA was extracted from stained fine needle aspirate slides (n = 95) or biopsy specimens (n = 15) taken during the initial diagnostic work up. The study protocol was approved by the Ethical Committee of the University of Ulm, and informed consent was obtained from all patients or volunteers.
DNA preparation. Plasma and serum was separated from peripheral blood collected in standard diagnostic tubes containing (or not) EDTA (1.6 mg/mL). Nucleated peripheral blood cells were isolated by ammonium chloride lysis of red blood cells. Cell pellets as well as serum and plasma samples were stored at −70°C. DNA was extracted from 200 μL serum or plasma, 0.1 to 2 × 107 peripheral blood cells, fine needle aspirate smears, or tumor biopsy specimens by proteinase K digestion, phenol-chloroform extraction,23 and ethanol precipitation. Glycogen (Boehringer Mannheim, Mannheim, Germany) was added at 20 μg/mL for precipitation of DNA extracted from serum, plasma, and fine needle aspirate smears. DNA was dissolved in standard Tris-EDTA buffer and stored at 4°C or −20°C.
PCR amplification of CDRIII DNA. For amplification of rearranged CDRIII DNA, a seminested PCR strategy was used: Thirty cycles of a first-round PCR with a consensus VH primer24 and a JH primer (no. 1 in Kneba et al25 ) were followed by 30 cycles using the same VH primer combined with a nested JH primer (no. 2 in Kneba et al25 ). Extensive screening of several primers described in the literature had previously shown that these primer combinations amplified 74% of a representative group of 196 lymphoma samples with maximal sensitivity and specificity (E. Müller and N. Frickhofen, unpublished results). One microliter of the first-round PCR product was used in the second-round PCR. PCR reactions were set up in a total volume of 30 μL and cycled in a Trio-Thermoblock (Biometra, Goettingen, Germany). Cloned Taq polymerase (Amplitaq; Perkin Elmer, Norwalk, CT) was used in a standard PCR buffer containing 3 mmol/L MgCl2 , 50 mmol/L KCl, 0.2 mmol/L deoxynucleotides and 15 pmol of each primer. Cycling conditions were identical in both PCR reactions: Initial denaturation for 120 seconds at 96°C was followed by 30 cycles of 60 seconds at 96°C, 60 seconds at 55°C, and 90 seconds at 72°C. Five to 12 μL of the final PCR product was run on an 8% nondenaturing polyacrylamide gel. DNA was visualized by ethidium bromide staining. A sample was defined as clonal, if at least two of three assays showed distinct bands of the same electrophoretic mobility. To show intact DNA, a 110-bp β-globin DNA fragment was amplified using primers GH20, PCO4, and PCO326 in a seminested PCR system similar to the one described previously.
Cloning and sequencing of tumor-derived CDRIII sequences. Selected PCR products were ligated into pGEM7Zf+, cloned in Escherichia coli DH5-α, sequenced using Sequenase V2.0 (United States Biochemicals, Cleveland, OH) or the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and a Model 373 or Model 310 DNA Sequencing System (Applied Biosystems).
Statistics. The rates of PCR-positive samples in patients with various clinical parameters of tumor load were compared by the two-tailed Fisher's exact test.
RESULTS
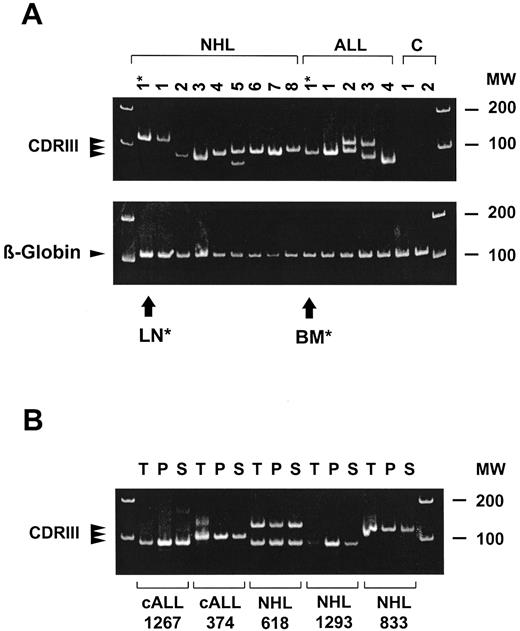
Prevalence of tumor-derived IgH DNA in serum or plasma of patients with B-cell malignancies. Clonal CDRIII PCR products were detected in at least one serum or plasma sample of 52 of 110 (47%) patients (see Fig 1A for representative CDRIII PCR amplifications). Sequencing showed identity of the CDRIII sequences amplified using DNA extracted from tumor cells, plasma, and serum in 10 of 10 randomly selected patients (Fig 1B and Table 1 show results from five representative patients). Tumor cells of 14 patients (12 patients with NHL and 2 patients with ALL) failed to be informative by the CDRIII-PCR technique used. In line herewith, none of the serum or plasma samples of these patients proved PCR positive. These patients excluded, the prevalence of patients with PCR-positive serum or plasma samples at any stage during the course of their treatment was 52 of 96 (54%). Using the criteria as described in Materials and Methods, none of the control serum or plasma samples showed a clonal CDRIII PCR product.
Detection of CDRIII DNA in the plasma of patients with B-cell malignancies. (A) CDRIII DNA amplified from the plasma of eight representative patients with NHL, four patients with B-precursor ALL at diagnosis, and two healthy control subjects. PCR-products derived from diagnostic tumor material of one NHL patient (lymph node: LN*, lane 1) and one ALL patient (bone marrow: BM*, lane 10) are included for comparison. Positive β-globin bands show the presence of DNA in all samples. (B) Comparison of CDRIII DNA amplification products in tumor cell samples (T), plasma (P), and serum (S) from five representative patients (two ALL and three NHL). PCR products were cloned and DNA sequences were found to be identical in each patient (summarized in Table 1). Ethidium bromide stained polyacrylamide gels; MW indicates molecular weight markers.
Detection of CDRIII DNA in the plasma of patients with B-cell malignancies. (A) CDRIII DNA amplified from the plasma of eight representative patients with NHL, four patients with B-precursor ALL at diagnosis, and two healthy control subjects. PCR-products derived from diagnostic tumor material of one NHL patient (lymph node: LN*, lane 1) and one ALL patient (bone marrow: BM*, lane 10) are included for comparison. Positive β-globin bands show the presence of DNA in all samples. (B) Comparison of CDRIII DNA amplification products in tumor cell samples (T), plasma (P), and serum (S) from five representative patients (two ALL and three NHL). PCR products were cloned and DNA sequences were found to be identical in each patient (summarized in Table 1). Ethidium bromide stained polyacrylamide gels; MW indicates molecular weight markers.
Clone-Specific CDRIII DNA Sequences of Representative ALL and NHL Patients
| . | Patient . | VH . | N-DH-N . | JH . |
|---|---|---|---|---|
| 1. | cALL 1267 | |||
| Tumor | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| Plasma | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| Serum | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| 2. | cALL 374 | |||
| Tumor | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| Plasma | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| Serum | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| 3a. | NHL 618 a | |||
| Tumor | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| Plasma | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| Serum | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| 3b. | NHL 618 b | |||
| Tumor | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| Plasma | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| Serum | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| 4. | NHL 1293 | |||
| Tumor | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| Plasma | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| Serum | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| 5. | NHL 833 | |||
| Tumor | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT | |
| Plasma | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT | |
| Serum | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT |
| . | Patient . | VH . | N-DH-N . | JH . |
|---|---|---|---|---|
| 1. | cALL 1267 | |||
| Tumor | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| Plasma | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| Serum | CGA | TTCGTTCATTACTATGGTTCGGGGAGTTAAC | CTA | |
| 2. | cALL 374 | |||
| Tumor | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| Plasma | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| Serum | CGAGAGA | TCGAAGATAGAGAGAGATAGCAGCTCGTCCTAC | TTGACTA | |
| 3a. | NHL 618 a | |||
| Tumor | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| Plasma | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| Serum | CGA | CGCCGCTTATAAGGGAAAAGGATATTGTGGTGGTGACTGCTATATCTGAGGC | GCTTTTGATAT | |
| 3b. | NHL 618 b | |||
| Tumor | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| Plasma | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| Serum | CGAGA | CAAAATAGTGCTTATTACCACGGG | GACTT | |
| 4. | NHL 1293 | |||
| Tumor | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| Plasma | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| Serum | CGAGAG | TAGATGGAAATCGCCGAGGCGTG | GACTA | |
| 5. | NHL 833 | |||
| Tumor | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT | |
| Plasma | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT | |
| Serum | CGAGAGA | CTTGGCTGTACCAGCTGGTATGCCCTTTTACAACTCTC | ACTACGGTATGGACGT |
CDRIII sequences in tumor cells, plasma, and serum of patients with NHL and ALL. Patients are identical to those in Fig 1B. In patient no. 618 two clonal rearrangements were shown.
Technical parameters influencing detection of tumor-derived IgH DNA. Because clonal DNA could be shown in only 54% of the samples tested, we were interested in determining whether this was a biological or technical finding. To address this question several variables possibly influencing the ability to detect clonal DNA in serum or plasma samples in vitro and in vivo were investigated.
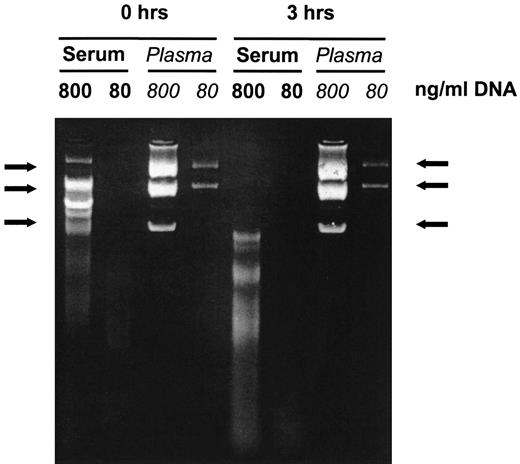
Clonotypic DNA could only be amplified from 24 of 87 (28%) of archival serum samples previously stored for up to 3 years in a serum bank of a diagnostic laboratory suggesting a major impact of the quality of serum on the success rate of the PCR. Spiking fresh blood samples of a healthy volunteer with plasmid DNA (pGEM7Zf+) showed that DNA degradation commences immediately on contact with blood. Addition of EDTA completely prevents DNA degradation as visualized by electrophoresis (Fig 2). This was confirmed after storage of plasmid DNA in whole blood for up to 5 days at room temperature. Similarly, 7 of 7 fresh, nonanticoagulated whole blood samples known to contain clonal IgH DNA remained PCR positive for 6 hours at room temperature and for 24 hours at 4°C, but all turned negative when stored for >36 hours (not shown). In contrast, blood drawn into EDTA remained PCR positive for at least 4 days at room temperature. β-globin sequences, as a marker of genomic DNA, could be amplified from all fresh serum samples and from 92% of the plasma samples stored for 2 days at room temperature.
Stability of DNA in plasma and serum. Blood of a normal volunteer was taken into tubes, previously spiked with plasmid DNA (pGEM7Zf+) to final concentrations as indicated. Serum or plasma was isolated immediately or after 3 hours of storage at room temperature. DNA was extracted, run on a 2% agarose gel, and stained with ethidium bromide. DNA in plasma is stable whereas DNA in serum is rapidly degraded. Arrows indicate intact plasmid DNA bands.
Stability of DNA in plasma and serum. Blood of a normal volunteer was taken into tubes, previously spiked with plasmid DNA (pGEM7Zf+) to final concentrations as indicated. Serum or plasma was isolated immediately or after 3 hours of storage at room temperature. DNA was extracted, run on a 2% agarose gel, and stained with ethidium bromide. DNA in plasma is stable whereas DNA in serum is rapidly degraded. Arrows indicate intact plasmid DNA bands.
Loss of DNA during extraction from serum or plasma might be another reason for false-negative PCR results. High concentrations of protein are a known cause of poor recovery of DNA by standard phenol-chloroform extraction methods. In line herewith, only 4 of 16 plasma samples from patients with myeloma were found to be positive by PCR. Two of the positive samples were obtained from patients with nonsecretory myeloma, and protein concentrations were less than 100 g/L in the other two patients (not shown). Extraction methods based on adsorption of DNA to matrices did not result in a higher yield of PCR-positive samples. Clonotypic PCR products could be amplified from both plasma and peripheral blood cells from 18 patients (13 NHL and 5 ALL), which could indicate that soluble DNA is released from tumor cells in vitro. However, PCR was positive in plasma but not in peripheral blood cells of another 6 patients (5 NHL and 1 ALL).
Clinical variables influencing detection of tumor-derived IgH DNA. Detection of tumor-derived DNA in the serum or plasma may also depend on tumor mass. However, median lactic dehydrogenase (LDH) concentrations were comparable in PCR-positive and PCR-negative patients investigated at diagnosis (317 v 389 U/L, P = .8) and there was only a trend for clinical parameters of tumor mass: five of 5 patients with bulky NHL and 10 of 11 patients with stage IV NHL at diagnosis were PCR positive compared with 16 of 21 patients with nonbulky disease (P = .5) and 11 of 15 patients with stage I to III disease (P = .4).
To investigate the effect of treatment on the ability to detect tumor cell–derived DNA in serum or plasma, blood was drawn at short intervals after commencement of treatment in three patients with ALL and two patients with NHL who reached clinical complete remission after induction treatment (daunorubicin, vincristin, and prednisolone for ALL and cyclophosphamide, doxorubicin, vincristin, and prednisone [CHOP] for NHL). In all patients serum and plasma samples remained PCR positive for 2 days after initiation of treatment but turned negative by day 5. Serial dilutions showed that there was no detectable increase in the total amount of circulating DNA immediately after start of treatment. The amount of DNA rather decreased steadily starting within 24 hours after the first infusion of cytotoxic drugs. In three of these patients (two ALL and one NHL), serum or plasma turned PCR positive again within 6 days before the next treatment cycle.
Because treatment obviously exerted profound effects on circulating DNA, PCR results were separately evaluated for patients who were investigated at diagnosis and those whose blood was drawn during or after chemotherapy (ie, more than 2 days after start and up to 2 weeks after completion of chemotherapy). About twice as many samples from patients at diagnosis were PCR positive compared with patients on or after treatment (Table 2).
Prevalence of Clonal CDRIII DNA in Cell-Free Blood Samples From Patients With NHL and ALL at Diagnosis and During Treatment
| Time of Sample Collection . | NHL . | ALL . | Total . |
|---|---|---|---|
| At diagnosis (n = 36) | |||
| Clonal CDRIII product | 21 | 10 | 31 (86%) |
| No clonal CDRIII product | 5 | 0 | 5 (14%) |
| On or after treatment (n = 60) | |||
| Clonal CDRIII product | 13 | 12 | 25 (42%) |
| No clonal CDRIII product | 24 | 11 | 35 (58%) |
| Time of Sample Collection . | NHL . | ALL . | Total . |
|---|---|---|---|
| At diagnosis (n = 36) | |||
| Clonal CDRIII product | 21 | 10 | 31 (86%) |
| No clonal CDRIII product | 5 | 0 | 5 (14%) |
| On or after treatment (n = 60) | |||
| Clonal CDRIII product | 13 | 12 | 25 (42%) |
| No clonal CDRIII product | 24 | 11 | 35 (58%) |
Only patients who were informative by PCR for tumor-derived CDRIII DNA are shown. Serum or plasma of patients was either sampled at the time of diagnosis or samples were taken at various times during or after treatment.
Correlation of cell-free clonotypic DNA with the clinical course. Persistence or disappearance of clonotypic DNA in serum or plasma may correlate with treatment results. Figure 3A illustrates a patient with mantle cell lymphoma who reached complete clinical remission after two courses of CHOP chemotherapy. However, she remained PCR positive in the plasma and relapsed 2 months later. In a patient with centroblastic lymphoma, clonotypic plasma DNA remained positive during CHOP chemotherapy. Plasma turned negative after induction of complete remission by high-dose chemotherapy and remained negative during further follow-up.
Clinical course and follow-up of clonotypic CDRIII DNA in the plasma of a patient with mantle cell lymphoma (patient no. 1,293, A) and a patient with centroblastic lymphoma (patient no. 581, B). The clinical course is indicated as active disease (solid bar), partial remission (grey bar), and complete remission (open bar). Treatment periods are indicated by boxes (CHOP, high-dose BCNU, etoposide, cytarabine, and melphalan [BEAM], followed by autologous peripheral blood stem cell transplantation, and methotrexate [MTX] and prednisone). Plasma was analyzed at diagnosis (DX ) and on five occasions during follow-up, given in weeks. Clinical response to high-dose chemotherapy, but not to standard CHOP treatment is paralleled by loss of detectable CDRIII DNA in the plasma. MW indicates molecular weight markers.
Clinical course and follow-up of clonotypic CDRIII DNA in the plasma of a patient with mantle cell lymphoma (patient no. 1,293, A) and a patient with centroblastic lymphoma (patient no. 581, B). The clinical course is indicated as active disease (solid bar), partial remission (grey bar), and complete remission (open bar). Treatment periods are indicated by boxes (CHOP, high-dose BCNU, etoposide, cytarabine, and melphalan [BEAM], followed by autologous peripheral blood stem cell transplantation, and methotrexate [MTX] and prednisone). Plasma was analyzed at diagnosis (DX ) and on five occasions during follow-up, given in weeks. Clinical response to high-dose chemotherapy, but not to standard CHOP treatment is paralleled by loss of detectable CDRIII DNA in the plasma. MW indicates molecular weight markers.
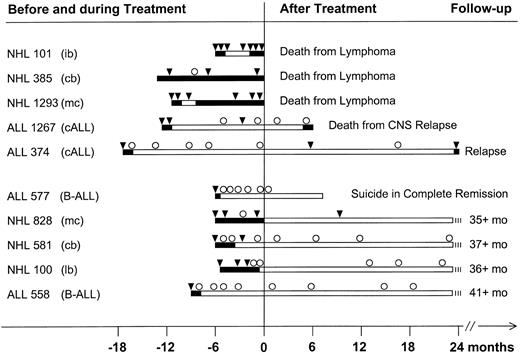
Longer molecular follow-up of 10 patients with NHL and ALL is illustrated in Fig 4. Several patterns emerge from these data: Patients whose serum or plasma was repeatedly positive for tumor-associated DNA (patients no. 101, 385, and 1,293) never reached remission, or remissions were short and they all eventually died from their disease. Occasional negative sera were drawn during chemotherapy and, thus, were not informative retrospectively (see previous data). In two patients with ALL, complete remission was reached by standard induction chemotherapy. In parallel their serum or plasma turned PCR negative (patients no. 1,267 and 374). In both patients there was an occasional DNA-positive serum or plasma (in both cases confirmed by extraction of separate samples to exclude contamination) among mostly negative samples. Patient no. 1,267 had an isolated central nervous system (CNS) relapse; her serum was still negative for CDRIII-DNA at that time. Patient no. 374 relapsed 25 months after the end of consolidation treatment. The last sample was drawn 9 months before clinical relapse and was PCR negative. Finally, serum or plasma of most patients whose disease did not relapse was consistently PCR negative (Fig 4, lower panel). The only exception was for patient no. 828: Despite being in complete clinical remission for more than 2.5 years, her serum contained clonotypic DNA 9 months before her last visit to the clinic. Because she suffered from mantle cell lymphoma, this seemingly favorable course must be viewed cautiously.
Long-term follow-up of clonotypic CDRIII-DNA in serum or plasma. Data from patients with NHL (n = 5) and ALL (n = 5) who did or did not relapse (5 patients each in the upper and lower panel). The patients shown in Fig 3 are included with longer follow-up. Clinically active disease and partial remission are depicted as solid bars and complete clinical remission is indicated by open bars. Molecular follow-up is shown above the clinical data: Serum or plasma was either positive (▾) or negative (○) for clonotypic CDRIII-DNA by PCR. ib, immunoblastic lymphoma; cb, centroblasic lymphoma; mc, mantle cell lymphoma; cALL, common ALL; B-ALL, surface immunoglobulin-positive B-cell ALL; lb, lymphoblastic lymphoma.
Long-term follow-up of clonotypic CDRIII-DNA in serum or plasma. Data from patients with NHL (n = 5) and ALL (n = 5) who did or did not relapse (5 patients each in the upper and lower panel). The patients shown in Fig 3 are included with longer follow-up. Clinically active disease and partial remission are depicted as solid bars and complete clinical remission is indicated by open bars. Molecular follow-up is shown above the clinical data: Serum or plasma was either positive (▾) or negative (○) for clonotypic CDRIII-DNA by PCR. ib, immunoblastic lymphoma; cb, centroblasic lymphoma; mc, mantle cell lymphoma; cALL, common ALL; B-ALL, surface immunoglobulin-positive B-cell ALL; lb, lymphoblastic lymphoma.
DISCUSSION
The study presented here indicates that tumor-derived DNA can be frequently recovered from serum and plasma of patients with B-cell leukemia or lymphoma whose tumor cells are informative on PCR analysis. Amplification of clonotypic DNA from serum or plasma, but not from leukocytes of several patients, and disappearance of amplifiable DNA from blood samples on storage suggested that this DNA is not released in vitro but rather circulates in vivo. DNA released from tumor cells in vitro may of course contribute to the DNA detected in whole blood samples of leukemic patients.
To our knowledge, detection of tumor-derived DNA in serum or plasma of patients with lymphoma or leukemia has not been reported before. However, there are reports of successful amplification of genomic or tumor-associated DNA from fresh or archival serum or plasma samples, supporting our observation.27-29 Similar to these investigators, we were able to amplify genomic DNA such as β-globin or p53 sequences from 100% of fresh serum and plasma samples. Even mRNA transcribed from housekeeping genes, such as glyceraldehyde-3-phosphate-dehydrogenase or tumor-specific hybrid RNA such as bcr-abl message, can be amplified from plasma as long as fresh samples are used (N. Frickhofen, unpublished results). Storage decreases the rate of positive serum samples because of degradation of DNA and RNA by nucleases in the serum. Addition of EDTA effectively blocks this degradation. DNA in whole blood samples anticoagulated with EDTA is stable for at least 4 days at room temperature. However, we prefer fresh serum over plasma because there were some PCR-negative plasma samples whereas serum drawn at the same time was PCR positive.
Is the detection of tumor-derived DNA in serum or plasma of leukemia and lymphoma patients more than a curiosity? First, our laboratory now routinely uses serum or plasma as a source of tumor-derived DNA if cells are not available. Serum or plasma can often be retrieved from freezers of clinical laboratories, whereas cells are rarely stored unless a biopsy has been performed. This holds particularly true for follow-up studies, because follow-up biopsies are rarely justified. The high success rate with genomic DNA may be also attractive for epidemiological studies using serum banks for population screening to detect hereditary germline mutations predisposing to cancer such as BRCA-1 in breast cancer, nonmalignant diseases such as CFTR in cystic fibrosis, or for demonstration of genomic instability in cancer patients.30 31
Second, serum and plasma complement blood and tissues as sources of tumor-derived DNA. There were several patients with stage I high-grade lymphoma whose serum or plasma was positive whereas their peripheral blood cells tested negative. In some of these patients, PCR with more sensitive clone-specific primers confirmed negative results in peripheral blood cells. This observation suggests that tumors that do not readily metastasize, may not shed many cells into the circulation but, nonetheless, release enough soluble DNA to be detected in serum or plasma.
Third, our follow-up results suggested that the clinical significance of serum- or plasma-derived DNA is different from that of DNA derived from peripheral blood cells, bone marrow, or other tissues. Disappearance of amplifiable DNA from serum and plasma within days after start of chemotherapy is intriguing. We expected an increase of DNA after destruction of tumor cells, similar to what can be observed with tumor-associated proteins. One explanation for this phenomenon would be that there is a steady state of DNA release from cells and degradation by plasma nucleases. Interference with DNA replication may shift this balance towards degradation of DNA. Thus, disappearance of circulating DNA could be an early indicator for successful chemotherapy in the sense of effective interference with DNA replication. If circulating tumor-derived DNA correlates with the presence of cells capable of DNA replication, a major problem of current PCR studies for minimal residual disease (that is, the question of whether this DNA is derived from proliferating tumor cells or from terminally differentiated cells or even cell debris) would be less critical.32
An important question to be answered is, What is the sensitivity of serum and plasma DNA as disease markers? Our results suggested that a positive PCR result is probably highly predictive for resistance to treatment and impending relapse. However, even when excluding methodological pitfalls such as poor serum quality and interference by chemotherapy, there are false-negative PCR results, because 14% of the serum or plasma samples of this series were negative by PCR at diagnosis and because there were also patients with negative follow-up studies who later relapsed. The rate of false-negative PCR studies may be higher for serum- and plasma-derived DNA than for cell-derived DNA because of the presence of nucleases in plasma with access to soluble DNA but not to cellular DNA. Thus, we consider the positive predictive value and the close correlation with active tumor as the major strength of this system.
In conclusion, DNA released from tumor cells into peripheral blood should be added to the list of markers that can be used to monitor cancer patients. Tumor cell DNA has been detected in the urine of patients with bladder cancer,33 in the pancreatic fluid of patients with pancreatic adenocarcinoma,34 and in the stool of patients with colon cancer.35 The use of unconventional sources of DNA could open new strategies for monitoring of cancer patients and may also prove useful for molecular epidemiology.36 The observation that naked DNA can be taken up and expressed by cells in vivo37 also brings to mind the possibility that soluble tumor-derived DNA could enter normal cells such as lymphocytes and macrophages and modify their function. It remains to be studied if soluble tumor-derived DNA can cause biological effects such as interference with spontaneous antidiotype responses or response to DNA vaccines.38-40 After all, nature must have had a reason to eliminate DNA and RNA in body fluids by means of potent nucleases.
ACKNOWLEDGMENT
We acknowledge the cooperation of all patients, nurses, and physicians who contributed to the collection of samples for this study. Prof T. Mertens kindly provided serum samples from the diagnostic serum bank of the Department of Virology, University of Ulm.
Supported by Deutsche Krebshilfe, Grant No. W61/91/Fr1 (to N.F.).
Address reprint requests to N. Frickhofen, MD, Dr-Horst-Schmidt-Kliniken, Department of Medicine III, Ludwig-Erhard-Str 100, D-65199 Wiesbaden, Germany.

![Fig. 3. Clinical course and follow-up of clonotypic CDRIII DNA in the plasma of a patient with mantle cell lymphoma (patient no. 1,293, A) and a patient with centroblastic lymphoma (patient no. 581, B). The clinical course is indicated as active disease (solid bar), partial remission (grey bar), and complete remission (open bar). Treatment periods are indicated by boxes (CHOP, high-dose BCNU, etoposide, cytarabine, and melphalan [BEAM], followed by autologous peripheral blood stem cell transplantation, and methotrexate [MTX] and prednisone). Plasma was analyzed at diagnosis (DX ) and on five occasions during follow-up, given in weeks. Clinical response to high-dose chemotherapy, but not to standard CHOP treatment is paralleled by loss of detectable CDRIII DNA in the plasma. MW indicates molecular weight markers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/12/10.1182_blood.v90.12.4953/3/m_bl_0025f3.jpeg?Expires=1769097778&Signature=fC4-g3RVWuxoVQBCNR0E~VlzNm8vZToh6ADTK1zVJ2S3a5Nt5xEhLEx3uPVewV-JJYIYaLrrza-uW5v8f3h~IA18rn00rvpfwWreylW9JKy1-w1HU7KfGth5ZkDW4BI69QVRlsYxZl5eZsmPxxAFh3t6Xz0vnaj9P3XrpljWGp0t8m3ilGWED5jiL6RfUirw2nl7bE9-rSOGM8K9zm7YgWDPs2Pie-9iDIbDAv0110AVmQRPHn2WQAMrRA2xXm1udEZ8Eh6kuL1XsbOyEKcHwe0SSmUssBn0NxoR6tJIzzHNG~YuytZexe29v08cpM6Vfc5sPtnXAzhz~0sM-V16SQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal