Abstract
Chromosome rearrangement of 14q32.33 has recurrently occurred with variable partner sites, including 11q13.3, 8q24.1, 18q21.3, and 6p21.1 in multiple myeloma (MM). To assess the actual incidence of 14q32.33 translocation and to elucidate its implication in the pathogenesis of MM, we studied 42 patients with MM, plasma cell leukemia, or plasmacytoma and 5 with monoclonal gammopathy with undetermined significance (MGUS) by G-banding and molecular cytogenetic methods. Using double-color fluorescence in situ hybridization (DCFISH) with 2 Ig heavy chain (IgH) gene probes, a yeast artificial chromosome (YAC) clone containing variable region, and a phage clone containing γ constant region, 14q32.33 translocation was detected as split signals of the IgH gene in 31 patients with plasma cell malignancies and 3 with MGUS. In contrast, of 40 patients who were assessed by G-banding, 3 (7.5%) showed the 14q+ chromosome. DCFISH detected a split of the IgH gene on interphase nuclei in 34 (73.9%) of 46 patients analyzed, whereas on metaphase spreads, it was in 22 (51.2%) of 43 patients analyzed. Interphase DCFISH was particularly useful to detect 14q32.33 translocation in 17 (65.4%) of 26 patients with normal karyotypes. Donor sites were identified in 11 of 22 patients demonstrated as carrying 14q32.33 translocation by metaphase FISH. Chromosome t(11; 14)(q13.3; q32.33) was detected in 5 patients, t(8; 14)(q24.1; q32.33) in 2, t(14; 18)(q32.33; q21.3) in 2, and t(7; 14)(q32.1; q32.33) in 1. A complex 14q32.33 translocation involving 3q and 16q24 was detected in 1 patient. Myeloma cells with t(7; 14) showed myelomonocytoid surface antigen. Because rearrangements of 14q32.33 were closely associated with translocation of proto-oncogenes into the IgH gene, our findings indicate that 14q32.33 translocation with various partner chromosomes is a critical event in the pathogenesis of MM and MGUS.
SPECIFIC CHROMOSOME abnormalities still remain to be elucidated in multiple myeloma (MM) and related disorders. Previous cytogenetic studies have shown that the incidence of numerical and structural rearrangement of chromosomes ranged from 20% to 60%.1-18 Although recent molecular cytogenetic studies have detected chromosomal aneuploidy in 90% of patients with MM,19,20 limited data were available for identifying recurrent chromosome rearrangements. We have previously shown that 14q32.33 translocation occurred in approximately 60% of cytogenetically abnormal cases.17,18 More recently, in MM cell lines, promiscuous translocations into the Ig heavy chain (IgH) gene locus have been reported by Bergsagel et al.21 However, the actual incidence of this cytogenetic alteration in MM remains unknown because of the low proliferative activity of myeloma cells. Correlations of cytogenetic findings with clinical features are a point of controversy as well.
On the other hand, in B-cell non-Hodgkin's lymphoma (NHL), 14q32.33 translocations are highly associated with histologic subtypes: t(8; 14)(q24.1; q32.33) with Burkitt's lymphoma, t(14; 18)(q32.33; q21.3) with follicular lymphoma, t(11; 14)(q13.3; q32.33) with mantle cell lymphoma, t(3; 14)(q27; q32.33) with large cell lymphoma, and t(9; 14) (p13; q32.33) with plasmacytoid lymphoma.22,23 For more than 10 years, proto-oncogenes or putative oncogenes were assigned to these loci: c-MYC to 8q24.1, BCL1 to 11q13.3, BCL2 to 18q21.3, BCL6 to 3q27, and PAX-5 to 9p13.24-33 The IgH gene translocates and juxtaposes these loci, resulting in dysregulation of respective oncogenes. Based on the site of the breakpoint within the IgH gene, it is postulated that the translocations are mediated by either aberrant VDJ or isotype switch recombination.21,24,28,34 In particular, translocations into the IgH gene locus occur preferentially in switch regions in MM cell lines.21,24 Among characteristic 14q32.33 translocations, 11q13, 8q24, 6p21.1, and 4p16.3 were recurrently reported as translocation partners in MM.1,3-7,9,10,12-19,21 35-37 Hence, molecular characterization of 14q32.33 translocation with a specific donor chromosome will provide significant information on the positional cloning of oncogenes closely associated with B-cell malignancies including MM.
Fluorescence in situ hybridization (FISH) technique has the advantage to detect the genetic defects in both metaphase spreads and interphase nuclei. Interphase FISH is particularly useful to detect the chromosomal rearrangements in cells of most differentiated malignancies in which no metaphase was available for precise analysis of the karyotype. Recently, multicolor FISH analysis has been established to simultaneously analyze multiple genetic rearrangements.38-40 In this respect, yeast artificial chromosome (YAC) clones are ideal FISH probes, because they yield excellent hybridization and signals after Alu-polymerase chain reaction (PCR) amplification of human-specific sequences.41,42 Using YAC clones, we have detected tumor-specific rearrangement of the IgH gene and BCL2 rearrangement in interphase nuclei.43 44
In the present study, to assess the actual incidence of 14q32.33 translocation and to elucidate its pathogenetic implication in MM and related disorders, we studied 47 patients with these diseases using double-color FISH (DCFISH) with the IgH gene probes and the conventional banding method. Chromosome 14q32.33 translocations were detected as split signals of the IgH gene in 31 of 42 patients with MM and plasma cell malignancies and 3 of 5 patients with monoclonal gammopathy of undetermined significance (MGUS).
MATERIALS AND METHODS
Patients.We studied 47 patients with plasma cell dyscrasias characterized clinically, hematologically, and immunologically; 38 patients had MM, 3 had plasma cell leukemia (PCL), 1 had plasmacytoma, and 5 had MGUS. The diagnosis of PCL was based on plasmacytosis exceeding 2 × 109/L of plasma cells in peripheral blood (PB).45 Patients were diagnosed as having MGUS according to standard criteria (<3 g/dL of serum monoclonal protein; <10% plasma cells in the marrow; absence of lytic bone lesions; and no anemia, hypercalcemia, or renal insufficiency).46 47 All procedures were performed for diagnostic purposes, and informed consent following institutional guidelines was obtained for using an aliquot of the specimens for research purposes. Ages of patients ranged from 39 to 83 years old; 28 were men and 19 were women. Types of the M-component were IgGκ in 24 patients, IgGλ in 13, BJκ in 3, BJλ in 2, IgAκ in 2, IgAλ in 2, and nonsecretary BJλ in 1. Myeloma cells in patient no. 11 expressed myelomonocytic surface markers; were positive for CD13, CD14, and CD33; but were negative for Sm-Ig, CD10, CD19, CD20, and glycoprotein IIb/IIIa. The selection of the patients was only based on the availability of preserved chromosome materials from January 1991 to December 1995.
Chromosome preparation and G-banding study.Chromosome preparation was performed by the short-term culture method.17 Briefly, bone marrow (BM), PB, and pleural effusion cells were cultured in RPMI 1640 medium supplemented with 15% bovine calf serum without any mitogen for 24 hours at 37°C in 5% CO2. To obtain elongated and finely banded chromosome, ethidium bromide (10 μg/mL) was added to the culture medium for 2 hours before treatment with colcemid (0.05 μg/mL) for 15 minutes. Cells were harvested with hypotonic potassium chloride and fixed by methanol/glacial acetic acid (3:1). Chromosomes were stained with Giemsa, pretreated by trypsin, and karyotyped according to the recommendation of the ISCN (1995).48
DCFISH.For DCFISH studies, we used the following probes: a YAC clone Y6 and a bacteriophage clone Igγ1-10 each containing variable region (VH)49 and Cγ1 region50 of the IgH gene locus, respectively. Cγ region is distanced approximately 1 Mb from sequences representative of Y6. These DNA sequences were mapped to the chromosome band 14q32.33. Our strategy was based on identifying the split and translocation of these sequences on metaphase and interphase nuclei. Human-specific DNA sequences were amplified from a YAC by Alu-PCR.42 Igγ1-10 was labeled by nick translation with biotin-16-dUTP (Boehringer Mannheim Biochemica, Mannheim, Germany). Alu-PCR products of Y6 were labeled with digoxigenin-11-dUTP (Boehringer Mannheim Biochemica). Mixed together, 100 ng of digoxigenin-labeled Y6 and 40 ng of biotin-labeled Igγ1-10 were used for suppression hybridization with 5 μg human Cot-1 DNA (GIBCO BRL, Grand Island, NY). A YAC clone yA153A6 containing the BCL2 gene (kindly provided by Gary A. Silverman, Harvard Medical School, Boston, MA) was used in combination with a VH gene probe to detect t(14; 18)(q32.33; q21.3). Signal detection was also performed as described previously.39 To detect donor sites of 14q32.33 translocations, chromosomes were identified using counterstaining with 4′,6-diaminido-2-phenylindole dihydrochloride (DAPI; 0.2μg/mL for 3 minutes). Control samples for interphase analysis were prepared from cytogenetically normal bone marrow cells obtained from 5 patients with mild leukocytosis or leukocytopenia. In all except for patient no. 31, interphase signals were evaluated in 50 to 200 nuclei (mainly 100 nuclei) per slides with a hybridization efficiency of more than 90%.
Metaphase and interphase cells were observed by fluorescence microscope (BX40-RF; Olympus, Tokyo, Japan). Double-band and triple-band-pass filter sets (Omega Optical, Brattleboro, VT) were used for simultaneously detecting fluorescein isothiocyanate (FITC), tetramethylrhodamine (TRITC), and DAPI fluorochromes. Photographs were taken with Fujichrome PROVIA1600 (Fuji film, Tokyo, Japan) or Kodak Ektachrome 400 color slide films (Kodak Inc, Rochester, NY). Slides were scanned for reproduction using a 35-mm slide scanner (Nikon Cool Scan II; Nikon, Tokyo, Japan) and edited using Adobe Photoshop, Version 3.0 (Adobe Systems, Mountain View, CA)
RESULTS
Delineation of the IgH gene translocation by DCFISH and controls.Two haptenized sequences of the IgH gene, VH and Cγ, were detected as yellow (Y) or connected red (R) and green (G) signals at chromosomal band 14q32.33 on normal metaphase spreads.39,44 Three signals each with yellow, green, and red color (YGR) on metaphase spreads and interphase nuclei typically indicated 14q32.33 translocation and a break between VH and Cγ3 regions (Fig 1a). The criteria for a positive IgH gene split in BM cells was according to the cut-off values (mean + 2 SD) as follows: 6.4% for YGR, 0% for YYG, 4.0% for YYR, 0% for GR, and 4.3% for YR. The criteria for a positive IgH gene split in peripheral blood cells was according to the cut-off values described previously.44
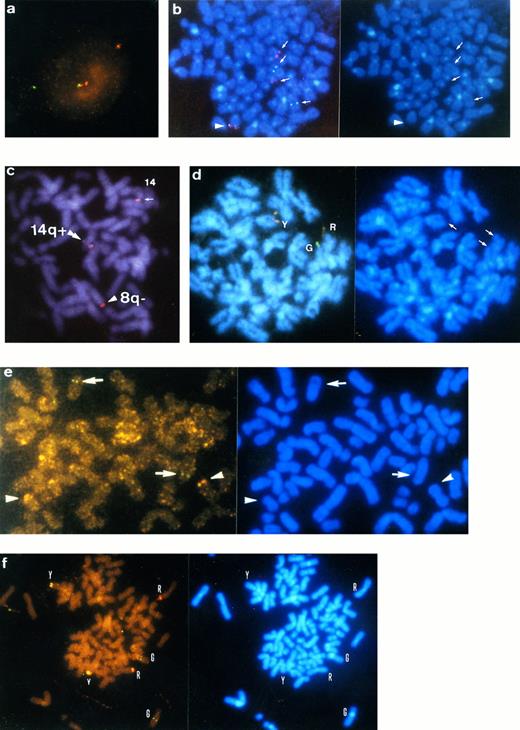
Interphase and metaphase FISH using VH and Cγ gene. DAPI banding pictures are shown on the right side of the corresponding FISH metaphases except for (c). (a) Split pattern of VH (TRITC, red) and Cγ (FITC, green) gene signals in interphase nuclei of patient no. 45 with MGUS. (b) Metaphase FISH on patient no. 41 with t(11; 14). Signals of VH (red) and Cγ (green) genes are observed on 11q− and 14q+, respectively (arrows). An arrowhead indicates normal chromosome 14. (c) Metaphase FISH on patient no. 1 with t(8; 14). Through tripple-band-pass filter, signals of split VH (arrowhead, pink) and Cγ-split VH (double arrowhead, whitish pink) genes are observed on 8q− and 14q+, respectively. Normal chromosome 14 was labeled with whitish pink signal of the IgH gene (arrow). In this patient, the breakpoint of the IgH gene is possibly within the variable region. (d) Metaphase FISH on patient no. 20 with t(14; 18). Cγ (G, green) and VH (R, red) signals are observed on 14q+ and 18q−, respectively. Y (yellow) denotes a normal allele of the IgH gene. (e) Metaphase FISH on patient no. 11 with t(7; 14). Signals of VH (arrowheads) and Cγ (arrows) genes are observed on 7q− and 14q+, respectively. (f ) Metaphase FISH on patient no. 2 with complex 14q32.33 translocation involving 3q and 16q24. Signals of VH (R) and Cγ (G) genes are observed on 3q− and 16q, respectively. Y indicates normal chromosome 14.
Interphase and metaphase FISH using VH and Cγ gene. DAPI banding pictures are shown on the right side of the corresponding FISH metaphases except for (c). (a) Split pattern of VH (TRITC, red) and Cγ (FITC, green) gene signals in interphase nuclei of patient no. 45 with MGUS. (b) Metaphase FISH on patient no. 41 with t(11; 14). Signals of VH (red) and Cγ (green) genes are observed on 11q− and 14q+, respectively (arrows). An arrowhead indicates normal chromosome 14. (c) Metaphase FISH on patient no. 1 with t(8; 14). Through tripple-band-pass filter, signals of split VH (arrowhead, pink) and Cγ-split VH (double arrowhead, whitish pink) genes are observed on 8q− and 14q+, respectively. Normal chromosome 14 was labeled with whitish pink signal of the IgH gene (arrow). In this patient, the breakpoint of the IgH gene is possibly within the variable region. (d) Metaphase FISH on patient no. 20 with t(14; 18). Cγ (G, green) and VH (R, red) signals are observed on 14q+ and 18q−, respectively. Y (yellow) denotes a normal allele of the IgH gene. (e) Metaphase FISH on patient no. 11 with t(7; 14). Signals of VH (arrowheads) and Cγ (arrows) genes are observed on 7q− and 14q+, respectively. (f ) Metaphase FISH on patient no. 2 with complex 14q32.33 translocation involving 3q and 16q24. Signals of VH (R) and Cγ (G) genes are observed on 3q− and 16q, respectively. Y indicates normal chromosome 14.
G-banding analysis.Forty patients were analyzed cytogenetically before FISH study as shown in Tables 1 and 2. Karyotypes of 2 patients (nos. 2 and 28) were described previously.18 Twenty-six patients showed normal karyotypes. Of 40 patients, 14 (35.0%) showed abnormal karyotypes. In 2 of these patients, an abnormal karyotype was presented as a single-cell abnormality (SCA; patients no. 9 and 15). Patients no. 14 and 44 also showed SCA with chromosomal aneuploidy. Karyotypic abnormalities were very complex, and the modal chromosomal number ranged from 42 to 78. Monosomy, trisomy, and tetrasomy were frequently observed as well as deletion, translocation, insertion, ring chromosome, isochromosome, and marker chromosome. Three patients (7.9%) had the 14q+ chromosome with an extrasegment derived from t(8; 14)(q24; q32) (patient no. 1) or from an unknown origin (patients no. 11 and 12). Two patients (nos. 2 and 31) showed t(8; 22)(q24; q11) (Fig 2).
FISH and Cytogenetic Analysis of MM and Related Disorders
| Patient No. . | Age/Sex . | M Protein . | Materials . | Plasma Cells . | G-Banded Karyotype* . | Metaphase FISH . | Interphase FISH . | Split Pattern‡ . | |
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | s/Total . | Donor† . | s/Total . | . |
| . | . | . | . | . | . | (%) . | . | (%) . | . |
| MM | |||||||||
| 1 | 65/F | A, λ | BM | 95.0 | t(8; 14) | 3/4 (75) | 8q24 | 89/100 (89) | YYR |
| 2 | 68/M | BJ, κ | BM | 90.8 | t(8; 22) | 6/7 (86) | 16q24 | 56/100 (56) | YGR |
| 3 | 72/F | BJ, κ | BM | 86.4 | 46,XX[25] | 0/25 (0) | 8/100 (8) | YGR | |
| 4 | 82/M | G, κ | BM | 71.2 | Abnormal | 0/22 (0) | 22/100 (22) | YGR | |
| 5 | 79/F | G, κ | BM | 60.8 | Abnormal | 4/20 (20) | Unknown | 16/100 (16) | YGR |
| 6 | 49/M | A, κ | BM | 54.0 | 46,XY[8] | 0/3 (0) | 12/100 (12) | YGR | |
| 7 | 55/M | G, λ | BM | 53.2 | 46,XY[8] | 0/8 (0) | 19/100 (19) | YR | |
| 8 | 83/F | G, κ | BM | 50.8 | 46,XX[2] | ND | 13/100 (13) | YGR | |
| 9 | 77/M | G, κ | BM | 45.6 | Abnormal | 2/16 (13) | Unknown | 25/100 (25) | YGR |
| 10 | 70/F | G, κ | BM | 44.4 | 46,XX[10] | 1/11 (9) | 11q13 | 38/100 (38) | YR |
| 11 | 42/M | BJ, λ | BM | 44.4 | 14q+ | 7/7 (100) | 7q32.1 | 60/100 (60) | GR |
| 12 | 50/M | G, λ | BM | 39.0 | 14q+ | 1/22 (5) | Unknown | 18/100 (18) | YGR |
| 13 | 54/M | A, λ | BM | 36.8 | ND | 1/6 (17) | Unknown | 24/100 (24) | YGR |
| 14 | 55/M | BJ, λ | BM | 34.2 | Abnormal | 0/10 (0) | 10/100 (10) | YGR | |
| 15 | 59/F | G, κ | BM | 32.0 | Abnormal | 0/17 (0) | 30/100 (30) | YGR | |
| 16 | 63/M | A, κ | BM | 30.0 | ND | 1/14 (7) | Unknown | 7/100 (7) | YGR |
| 17 | 68/M | G, κ | BM | 28.8 | 46,XY[4] | 0/6 (0) | 24/100 (24) | YGR | |
| 18 | 39/M | BJ, κ | BM | 26.0 | 46,XY[13] | 3/18 (17) | 8q24 | 22/100 (22) | YGR |
| 19 | 62/M | G, λ | BM | 24.0 | Abnormal | 5/18 (28) | Unknown | 24/200 (12) | YGR |
| 20 | 66/M | G, κ | BM | 21.6 | 46,XY[20] | 2/24 (8) | 18q21 | 19/100 (19) | YGR |
| 21 | 67/F | G, κ | BM | 19.6 | 46,XX[20] | 1/14 (7) | Unknown | 10/100 (10) | YGR |
| 22 | 65/M | G, λ | BM | 19.2 | 46,XY[5] | ND | 17/100 (17) | YGR | |
| 23 | 76/F | G, λ | BM | 16.0 | 46,XX[5] | 2/20 (10) | Unknown | 10/100 (10) | YGR |
| 25 | 63/M | G, κ | BM | 15.2 | 46,XY[10] | 2/21 (10) | Unknown | 14/100 (14) | YGR |
| 24 | 61/F | G, κ | BM | 15.0 | 46,XX[20] | 4/16 (25) | 18q21 | 17/100 (17) | YGR |
| 26 | 75/F | G, λ | BM | 13.8 | ND | 1/7 (14) | Unknown | 11/100 (11) | YGR |
| 27 | 44/F | G, κ | BM | 13.2 | 46,XX[20] | 0/31 (0) | 10/100 (10) | YGR | |
| 28 | 82/F | G, λ | PE | 88.1 | Abnormal | 0/11 (0) | 1/100 (1) | ||
| 29 | 70/F | G, κ | BM | 59.2 | 46,XX[7] | 0/8 (0) | 6/100 (6) | ||
| 30 | 62/F | G, κ | BM | 17.4 | 46,XX[3] | ND | 4/100 (4) | ||
| 40.6ρ | |||||||||
| 31 | 54/M | G, κ | BM | 44.0 | t(8; 22) | 0/5 (0) | Unevaluable1-155 | ||
| 32 | 75/M | G, λ | BM | 43.0 | ND | 0/10 (0) | 1/50 (2) | ||
| 33 | 73/F | G, κ | BM | 36.4 | ND | ND | 5/100 (5) | ||
| 34 | 74/M | G, κ | BM | 33.6 | 46,XY[20] | 0/20 | 1/100 (1) | ||
| 35 | 81/M | G, κ | BM | 20.8 | 46,XY[5] | 0/11 (0) | 4/100 (4) | ||
| 36 | 65/M | G, λ | BM | 15.8 | 46,XY[20] | 0/15 (0) | 5/100 (5) | ||
| 37 | 62/M | G, κ | BM | 15.6 | 46,XY[22] | 0/26 (0) | 3/100 (3) | ||
| 38 | 60/M | G, κ | BM | 12.0 | 46,XY[18] | 0/4 (0) | 4/100 (4) | ||
| Plasma cell leukemia | |||||||||
| 39 | 66/M | G, κ | BM | 84.4 | 46,XY[5] | 4/6 (67) | 11q13 | 55/100 (55) | YGR |
| 401-154 | 79/F | BJ, (nonsecretary) | PB | 80.0 | 11q− | 3/5 (60) | 11q13 | 13/100 (13) | YYGG |
| 41 | 62/M | G, κ | PB | 28.0 | ND | 2/3 (67) | 11q13 | 15/100 (15) | YGR |
| Plasmacytoma | |||||||||
| 42 | 71/F | G, λ | BM | 19.8 | 46,XX[20] | 1/12 (8) | 11q13 | 8/50 (16) | YGR |
| MGUS | |||||||||
| 43 | 73/M | G, κ | BM | 9.6 | ND | 0/10 (0) | 18/200 (9) | YGR | |
| 44 | 73/M | G, λ | BM | 5.6 | Abnormal | 2/13 (15) | Unknown | 11/100 (11) | YGR |
| 45 | 78/M | G, λ | BM | 4.8 | 46,XY[20] | 0/14 (0) | 8/100 (8) | YGR | |
| 46 | 71/F | G, λ | BM | 8.8 | 46,XX[12] | 0/24 (0) | 3/100 (3) | ||
| 47 | 80/F | G, κ | BM | 1.6 | 46,XX[20] | 0/30 (0) | 4/100 (4) | ||
| Patient No. . | Age/Sex . | M Protein . | Materials . | Plasma Cells . | G-Banded Karyotype* . | Metaphase FISH . | Interphase FISH . | Split Pattern‡ . | |
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | s/Total . | Donor† . | s/Total . | . |
| . | . | . | . | . | . | (%) . | . | (%) . | . |
| MM | |||||||||
| 1 | 65/F | A, λ | BM | 95.0 | t(8; 14) | 3/4 (75) | 8q24 | 89/100 (89) | YYR |
| 2 | 68/M | BJ, κ | BM | 90.8 | t(8; 22) | 6/7 (86) | 16q24 | 56/100 (56) | YGR |
| 3 | 72/F | BJ, κ | BM | 86.4 | 46,XX[25] | 0/25 (0) | 8/100 (8) | YGR | |
| 4 | 82/M | G, κ | BM | 71.2 | Abnormal | 0/22 (0) | 22/100 (22) | YGR | |
| 5 | 79/F | G, κ | BM | 60.8 | Abnormal | 4/20 (20) | Unknown | 16/100 (16) | YGR |
| 6 | 49/M | A, κ | BM | 54.0 | 46,XY[8] | 0/3 (0) | 12/100 (12) | YGR | |
| 7 | 55/M | G, λ | BM | 53.2 | 46,XY[8] | 0/8 (0) | 19/100 (19) | YR | |
| 8 | 83/F | G, κ | BM | 50.8 | 46,XX[2] | ND | 13/100 (13) | YGR | |
| 9 | 77/M | G, κ | BM | 45.6 | Abnormal | 2/16 (13) | Unknown | 25/100 (25) | YGR |
| 10 | 70/F | G, κ | BM | 44.4 | 46,XX[10] | 1/11 (9) | 11q13 | 38/100 (38) | YR |
| 11 | 42/M | BJ, λ | BM | 44.4 | 14q+ | 7/7 (100) | 7q32.1 | 60/100 (60) | GR |
| 12 | 50/M | G, λ | BM | 39.0 | 14q+ | 1/22 (5) | Unknown | 18/100 (18) | YGR |
| 13 | 54/M | A, λ | BM | 36.8 | ND | 1/6 (17) | Unknown | 24/100 (24) | YGR |
| 14 | 55/M | BJ, λ | BM | 34.2 | Abnormal | 0/10 (0) | 10/100 (10) | YGR | |
| 15 | 59/F | G, κ | BM | 32.0 | Abnormal | 0/17 (0) | 30/100 (30) | YGR | |
| 16 | 63/M | A, κ | BM | 30.0 | ND | 1/14 (7) | Unknown | 7/100 (7) | YGR |
| 17 | 68/M | G, κ | BM | 28.8 | 46,XY[4] | 0/6 (0) | 24/100 (24) | YGR | |
| 18 | 39/M | BJ, κ | BM | 26.0 | 46,XY[13] | 3/18 (17) | 8q24 | 22/100 (22) | YGR |
| 19 | 62/M | G, λ | BM | 24.0 | Abnormal | 5/18 (28) | Unknown | 24/200 (12) | YGR |
| 20 | 66/M | G, κ | BM | 21.6 | 46,XY[20] | 2/24 (8) | 18q21 | 19/100 (19) | YGR |
| 21 | 67/F | G, κ | BM | 19.6 | 46,XX[20] | 1/14 (7) | Unknown | 10/100 (10) | YGR |
| 22 | 65/M | G, λ | BM | 19.2 | 46,XY[5] | ND | 17/100 (17) | YGR | |
| 23 | 76/F | G, λ | BM | 16.0 | 46,XX[5] | 2/20 (10) | Unknown | 10/100 (10) | YGR |
| 25 | 63/M | G, κ | BM | 15.2 | 46,XY[10] | 2/21 (10) | Unknown | 14/100 (14) | YGR |
| 24 | 61/F | G, κ | BM | 15.0 | 46,XX[20] | 4/16 (25) | 18q21 | 17/100 (17) | YGR |
| 26 | 75/F | G, λ | BM | 13.8 | ND | 1/7 (14) | Unknown | 11/100 (11) | YGR |
| 27 | 44/F | G, κ | BM | 13.2 | 46,XX[20] | 0/31 (0) | 10/100 (10) | YGR | |
| 28 | 82/F | G, λ | PE | 88.1 | Abnormal | 0/11 (0) | 1/100 (1) | ||
| 29 | 70/F | G, κ | BM | 59.2 | 46,XX[7] | 0/8 (0) | 6/100 (6) | ||
| 30 | 62/F | G, κ | BM | 17.4 | 46,XX[3] | ND | 4/100 (4) | ||
| 40.6ρ | |||||||||
| 31 | 54/M | G, κ | BM | 44.0 | t(8; 22) | 0/5 (0) | Unevaluable1-155 | ||
| 32 | 75/M | G, λ | BM | 43.0 | ND | 0/10 (0) | 1/50 (2) | ||
| 33 | 73/F | G, κ | BM | 36.4 | ND | ND | 5/100 (5) | ||
| 34 | 74/M | G, κ | BM | 33.6 | 46,XY[20] | 0/20 | 1/100 (1) | ||
| 35 | 81/M | G, κ | BM | 20.8 | 46,XY[5] | 0/11 (0) | 4/100 (4) | ||
| 36 | 65/M | G, λ | BM | 15.8 | 46,XY[20] | 0/15 (0) | 5/100 (5) | ||
| 37 | 62/M | G, κ | BM | 15.6 | 46,XY[22] | 0/26 (0) | 3/100 (3) | ||
| 38 | 60/M | G, κ | BM | 12.0 | 46,XY[18] | 0/4 (0) | 4/100 (4) | ||
| Plasma cell leukemia | |||||||||
| 39 | 66/M | G, κ | BM | 84.4 | 46,XY[5] | 4/6 (67) | 11q13 | 55/100 (55) | YGR |
| 401-154 | 79/F | BJ, (nonsecretary) | PB | 80.0 | 11q− | 3/5 (60) | 11q13 | 13/100 (13) | YYGG |
| 41 | 62/M | G, κ | PB | 28.0 | ND | 2/3 (67) | 11q13 | 15/100 (15) | YGR |
| Plasmacytoma | |||||||||
| 42 | 71/F | G, λ | BM | 19.8 | 46,XX[20] | 1/12 (8) | 11q13 | 8/50 (16) | YGR |
| MGUS | |||||||||
| 43 | 73/M | G, κ | BM | 9.6 | ND | 0/10 (0) | 18/200 (9) | YGR | |
| 44 | 73/M | G, λ | BM | 5.6 | Abnormal | 2/13 (15) | Unknown | 11/100 (11) | YGR |
| 45 | 78/M | G, λ | BM | 4.8 | 46,XY[20] | 0/14 (0) | 8/100 (8) | YGR | |
| 46 | 71/F | G, λ | BM | 8.8 | 46,XX[12] | 0/24 (0) | 3/100 (3) | ||
| 47 | 80/F | G, κ | BM | 1.6 | 46,XX[20] | 0/30 (0) | 4/100 (4) | ||
Abbreviations: s, split; PE, pleural effusion; ND, not done.
A karyotype was described in Table 2.
Donor sites were identified on the basis of the DAPI banding pattern.
Representative pattern of split signal was described.
ρ Plasmacytoid lymphocyte.
Interphase nuclei were unevaluable because of background noise in spite of repeated experiments.
Patient no. 40 showed complicated split signal pattern with yellow signals on the normal chromosome 14 and on the 11q− and green signals on the two marker chromosomes (YYGG pattern). Based on these findings, we counted nuclei carrying YYGG pattern and presented the number of positive nuclei in the table.
Karyotypes in 14 Patients With Chromosomal Abnormalities
| Patient No. . | Karyotype . |
|---|---|
| 1 | 52,X,del(X)(p21p22),+i(1)(q10),t(4; 6)(q33;q21),?t(5; 20)(q35;q11),+7,del(7)(q22q32)×2,t(8; 14)(q24;q32),+9,?t(10; 15)(q11;q22),del(11) |
| (q13q21),i(12)(p10),der(12)t(12; 18)(q10;q10),+15,+17,+18,del(20)(q11)[14]/46,XX[5] | |
| 2* | 42,X,−Y,der(1)t(1; 1)(p13.3;q12),+3,t(8; 22)(q24;q11),i(9)(q10),−11,−13,−14,−15,add(16)(q24),add(19)(p13)[8] |
| 4 | 55,XY,+3,+5,+7,−8,+9,der(11)t(1; 11)(q21;p15),−13,−14,+15,+16,+17,+21×2,+3mar[2]/45,X,−Y[4]/46,XY[14] |
| 5 | 46,XX,del(6)(q14-16q23)[3]/46,XX[17] |
| 9 | 3n−1:63,XY,−2,+5,t(8; 16)(q24;q12),+11,−13,−13,−14,−16,del(16)(q22),−17,−18,−20,+21,−22,+r[1]/46,XY[18] |
| 11 | 81,XXY,−Y,−1×2,−4,−5,del(6)(q21q27)×2,del(7)(q32)×2,+del(7)(q22)×2,−8,inv(9)(p13;q13)×2,−13×2,add(14)(q32)×2,−14×2,−15, |
| −16×2,−17,−18×2,+der(19)t(1; 19)(q21;p13.3)×3,−21,−22,+marx2[12]/46,XY[8] | |
| 12 | 46,XY,7p+,8q+,14q+,16q+,17q+[cp8]/46,XY[8] |
| 14 | 46,XY,+3,+6,−13,−18[1]/46,XY[4] |
| 15 | 49,XX,add(1)(p10),−7,+8,+9,+11,+15,−16,+19[1]/46,XX[19] |
| 19 | 51,XY,+X,−1,+3p−,+5,6q−,+7q+,16,−18×2,+19,+20q+,+22,+mar[3]/46,XY[12] |
| 28* | 45,X,add(X)(q22),add(1)(p11),t(3; 17)(p21;p11),add(6)(q25),der(9)t(1; 9)(q11;p13),−13[3]/46,XX[7] |
| 31 | 59,XY,+del(2)(q21q31),+3,+del(5)(q12q15),der(7)t(1; 7)(q21;p22),t(8; 22)(q24;q11),+9×2,+11,+12,+15,+19,+21,+mar1×2,+mar2[7]/46,XY[9] |
| 40 | 42,X,add(X)(q28),t(1; 5)(p22;q33−35),+4,−6,add(7)(p22),−8,−10,+11,del(11)(q11),del(11)(q13),−13,−14,−17,−20,−22×2,+3mar[3] |
| 44 | 47,XY,+8[1]/46,XY[2] |
| Patient No. . | Karyotype . |
|---|---|
| 1 | 52,X,del(X)(p21p22),+i(1)(q10),t(4; 6)(q33;q21),?t(5; 20)(q35;q11),+7,del(7)(q22q32)×2,t(8; 14)(q24;q32),+9,?t(10; 15)(q11;q22),del(11) |
| (q13q21),i(12)(p10),der(12)t(12; 18)(q10;q10),+15,+17,+18,del(20)(q11)[14]/46,XX[5] | |
| 2* | 42,X,−Y,der(1)t(1; 1)(p13.3;q12),+3,t(8; 22)(q24;q11),i(9)(q10),−11,−13,−14,−15,add(16)(q24),add(19)(p13)[8] |
| 4 | 55,XY,+3,+5,+7,−8,+9,der(11)t(1; 11)(q21;p15),−13,−14,+15,+16,+17,+21×2,+3mar[2]/45,X,−Y[4]/46,XY[14] |
| 5 | 46,XX,del(6)(q14-16q23)[3]/46,XX[17] |
| 9 | 3n−1:63,XY,−2,+5,t(8; 16)(q24;q12),+11,−13,−13,−14,−16,del(16)(q22),−17,−18,−20,+21,−22,+r[1]/46,XY[18] |
| 11 | 81,XXY,−Y,−1×2,−4,−5,del(6)(q21q27)×2,del(7)(q32)×2,+del(7)(q22)×2,−8,inv(9)(p13;q13)×2,−13×2,add(14)(q32)×2,−14×2,−15, |
| −16×2,−17,−18×2,+der(19)t(1; 19)(q21;p13.3)×3,−21,−22,+marx2[12]/46,XY[8] | |
| 12 | 46,XY,7p+,8q+,14q+,16q+,17q+[cp8]/46,XY[8] |
| 14 | 46,XY,+3,+6,−13,−18[1]/46,XY[4] |
| 15 | 49,XX,add(1)(p10),−7,+8,+9,+11,+15,−16,+19[1]/46,XX[19] |
| 19 | 51,XY,+X,−1,+3p−,+5,6q−,+7q+,16,−18×2,+19,+20q+,+22,+mar[3]/46,XY[12] |
| 28* | 45,X,add(X)(q22),add(1)(p11),t(3; 17)(p21;p11),add(6)(q25),der(9)t(1; 9)(q11;p13),−13[3]/46,XX[7] |
| 31 | 59,XY,+del(2)(q21q31),+3,+del(5)(q12q15),der(7)t(1; 7)(q21;p22),t(8; 22)(q24;q11),+9×2,+11,+12,+15,+19,+21,+mar1×2,+mar2[7]/46,XY[9] |
| 40 | 42,X,add(X)(q28),t(1; 5)(p22;q33−35),+4,−6,add(7)(p22),−8,−10,+11,del(11)(q11),del(11)(q13),−13,−14,−17,−20,−22×2,+3mar[3] |
| 44 | 47,XY,+8[1]/46,XY[2] |
Cases previously reported.18
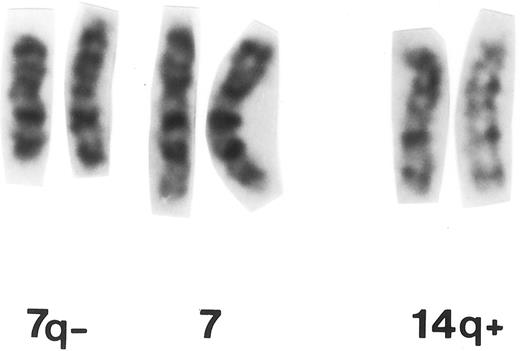
Partial G-banded karyotype of patient no. 11 with MM carrying t(7; 14)(q32.1; q32.33).
Partial G-banded karyotype of patient no. 11 with MM carrying t(7; 14)(q32.1; q32.33).
Among 14 patients with abnormal karyotype, 8 (57%) patients (no. 1, 2, 4, 11, 15, 28, 31, and 40) showed structural aberrations of chromosome 1. Rearrangements of chromosome 6q were found in 5 (36%) patients (nos. 1, 5, 11, 19, and 28). Chromosome 19p13 translocations were observed in 2 (17%) patients (nos. 2 and 11), each concomitantly with t(8; 22) or add(14)(q32.33).
Metaphase FISH analysis.Metaphase spreads were obtained from 43 of 47 patients for FISH studies, and 3 to 31 metaphases were evaluated per slides (Tables 1 and 3). Of 43 patients, 22 (51.1%) showed metaphase cells with split signals at a frequency of 5% to 100% (Tables 1 and 3). FISH identified the donor of 14q+ chromosomes in 11 patients based on the DAPI banding pattern of derivative chromosomes that were labeled with red or yellow signal (distal sequence of the IgH gene), invariably on the chromosome terminal band. Chromosome 14q+ was identified with green signal (proximal sequence of the IgH gene). As shown in Table 4, partner sites were 11q13 in 5 patients (nos. 10, 39, 40, 41, and 42; Fig 1b), 8q24 in 2 (nos. 1 and 18; Fig 1c) , 18q21 in 2 (nos. 20 and 24; Fig 1d), and 7q32.1 in 1 (no. 11; Fig 1e). Chromosome t(14; 18) was confirmed by demonstrating a fusion of VH and BCL2 genes with 2 YAC clones, Y6 and yA153A6. In patient no. 11, green and red signals were observed on 14q+ and on 7q−, respectively, whereas no yellow signal was observed (GR pattern; Fig 1e). Chromosomal band 16q24 was involved in a complex 14q32.33 translocation in patient no. 2 who was described in our previous report (Fig 1f ).18 VH (red) segment was observed on the terminus of the long arm of der(3)t(3; 14)(q25; q23) and Cγ (green) sequences at band 16q24 of der(16)t(15; 16) (q11; q24), which was identified based on the heteromorphism of chromosome 16, stained specifically with DAPI (Fig 1f ). A normal chromosome 14 was also observed with yellow signal on the terminal band. These findings suggest an insertion of Cγ into the breakpoint of the16q24 band on der(16)t(15; 16). Polyploid metaphase cells were also found; two sets of YGR signal were detected in 4 patients.
Comparison Between Metaphase and Interphase FISH
| Metaphase FISH . | No. of Patients . | Interphase FISH . | ||
|---|---|---|---|---|
| . | . | S(+) . | S(−) . | NI . |
| S(+) | 22 | 22 | 0 | 0 |
| S(−) | 21 | 10 | 10 | 1 |
| NI | 4 | 2 | 2 | 0 |
| Total | 47 | 34 (72.3%) | 12 | 1 |
| Metaphase FISH . | No. of Patients . | Interphase FISH . | ||
|---|---|---|---|---|
| . | . | S(+) . | S(−) . | NI . |
| S(+) | 22 | 22 | 0 | 0 |
| S(−) | 21 | 10 | 10 | 1 |
| NI | 4 | 2 | 2 | 0 |
| Total | 47 | 34 (72.3%) | 12 | 1 |
Abbreviations: S, split of signals of the IgH gene; NI, not informative.
Donor Sites of 14q32.33 Translocations Detected by FISH
| Specific Translocations . | Partner Sites . | Disease4-150 . | Patient No. . | Karyotypic Abnormalities . |
|---|---|---|---|---|
| t(11; 14) | 11q13 | MM (G, κ) | 10 | 46,XX |
| PCL (G, κ) | 39 | 46,XY | ||
| PCL (BJ, λ) | 40 | 11q− | ||
| PCL (G, κ) | 41 | ND | ||
| PCM (G, λ) | 42 | 46,XX | ||
| t(8; 14) | 8q24 | MM (A, λ) | 1 | t(8; 14) |
| MM (BJ, κ) | 18 | 46,XY | ||
| t(14; 18) | 18q21 | MM (G, κ) | 20 | 46,XY |
| MM (G, κ) | 24 | 46,XX | ||
| t(7; 14) | 7q32.1 | MM (BJ, λ)4-150 | 11 | 14q+ |
| ins[14;der(16)] | 16q24 | MM (BJ, κ) | 2 | t(8; 22) |
| t(14;?) | NI | MM (G, κ) | 5 | 6q− |
| MM (G, κ) | 9 | Complex | ||
| MM (G, λ) | 12 | 14q+ | ||
| MM (A, λ) | 13 | ND | ||
| MM (A, κ) | 16 | ND | ||
| MM (G, κ) | 19 | Complex | ||
| MM (G, κ) | 21 | 46,XY | ||
| MM (G, λ) | 23 | 46,XX | ||
| MM (G, κ) | 25 | 46,XY | ||
| MM (G, λ) | 26 | ND | ||
| MGUS (G, λ) | 44 | +8(SCA) |
| Specific Translocations . | Partner Sites . | Disease4-150 . | Patient No. . | Karyotypic Abnormalities . |
|---|---|---|---|---|
| t(11; 14) | 11q13 | MM (G, κ) | 10 | 46,XX |
| PCL (G, κ) | 39 | 46,XY | ||
| PCL (BJ, λ) | 40 | 11q− | ||
| PCL (G, κ) | 41 | ND | ||
| PCM (G, λ) | 42 | 46,XX | ||
| t(8; 14) | 8q24 | MM (A, λ) | 1 | t(8; 14) |
| MM (BJ, κ) | 18 | 46,XY | ||
| t(14; 18) | 18q21 | MM (G, κ) | 20 | 46,XY |
| MM (G, κ) | 24 | 46,XX | ||
| t(7; 14) | 7q32.1 | MM (BJ, λ)4-150 | 11 | 14q+ |
| ins[14;der(16)] | 16q24 | MM (BJ, κ) | 2 | t(8; 22) |
| t(14;?) | NI | MM (G, κ) | 5 | 6q− |
| MM (G, κ) | 9 | Complex | ||
| MM (G, λ) | 12 | 14q+ | ||
| MM (A, λ) | 13 | ND | ||
| MM (A, κ) | 16 | ND | ||
| MM (G, κ) | 19 | Complex | ||
| MM (G, κ) | 21 | 46,XY | ||
| MM (G, λ) | 23 | 46,XX | ||
| MM (G, κ) | 25 | 46,XY | ||
| MM (G, λ) | 26 | ND | ||
| MGUS (G, λ) | 44 | +8(SCA) |
Abbreviations: NI, not identified; PCL, plasma cell leukemia; PCM, plasmacytoma; SCA, single cell abnormality; ND, not done.
Myeloma cells showed myelomonocytoid phenotype.
Two patients with t(8; 14)(q24.1; q32.33) showed different split signal patterns of the IgH gene probes. In patient no. 18, red signal was at band 8q24.1 on the donor chromosome and a green signal was on the breakpoint of 14q32.33 juxtaposing 8q24.1 of the donor sequence, indicating that rearrangement occurred between Cγ and VH. On the other hand, in patient no. 1, abnormal YYR pattern consisted of one pink (corresponding to R) and two whitish pink signals (corresponding to Y) through triple-band pass filter (Fig 1c), suggesting translocation into VH region.
Interphase FISH analysis.Interphase FISH analysis was successfully performed in 46 of 47 patients. Intraclonal heterogeneity of signal patterns was commonly observed. In addition to YGR pattern, YG, YR, and GR with or without multiplication were also found in significant number of cells. These complex findings are possibly due to deletion of the IgH gene segments resulted from VDJ and isotype switch recombination occurring in myeloma cell before the 14q32.33 translocation or due to deletion of derivative chromosome. Polyploid or aneuploid nuclei were frequently encountered; two sets of YGR were detected in 5 patients. In 12 patients, 3 to 7 yellow signals were found, although it was unclear whether those signals originated from polysomy 14 of myeloma cells or normal megakaryocytes.
To analyze unequivocal data, YGR, YYG, YYR, GR, and YR patterns were evaluated, and figures of representative patterns are given in Table 1. Of 46 patients, 34 (73.9%) showed the IgH gene split. YGR pattern was observed in 29 patients, whereas YR was found in 2, and GR, YYR, and YYGG were each found in 1. The frequency of positive cells ranged from 7% to 89% in 34 patients according to the respective cut-off values.
Comparison between G-banding and FISH studies.Incidence of 14q32.33 translocation was shown according to the cytogenetic category in Table 5. In all 3 of the patients with 14q32.33 translocation assessed by G-banding, FISH analysis showed IgH translocation accordingly. Of 2 patients with t(8; 22), 1 showed IgH gene translocation. Interestingly, a cell line harboring t(8; 14) was established from this patient (unpublished data). Of 9 patients with cytogenetic aberrations other than 14q+ chromosome and its variant form, 8 showed IgH gene translocation.
Incidence of IgH Gene Translocation in 47 Patients With MM and Related Disorders
| Cytogenetic Category . | No. of Patients . | IgH Gene Translocation . | ||
|---|---|---|---|---|
| . | . | Interphase . | Metaphase . | Total . |
| 14q32 translocations | 3 | 3 | 3 | 3 |
| t(8; 22)(q24;q11) | 2 | 1 | 1 | 1 |
| Other abnormalities | 9 | 8 | 5 | 8 |
| Normal karyotype | 26 | 17 | 9 | 17 |
| Not done | 7 | 5 | 4 | 5 |
| Total | 47 | 34 (72.3%) | 22 (46.8%) | 34 |
| Cytogenetic Category . | No. of Patients . | IgH Gene Translocation . | ||
|---|---|---|---|---|
| . | . | Interphase . | Metaphase . | Total . |
| 14q32 translocations | 3 | 3 | 3 | 3 |
| t(8; 22)(q24;q11) | 2 | 1 | 1 | 1 |
| Other abnormalities | 9 | 8 | 5 | 8 |
| Normal karyotype | 26 | 17 | 9 | 17 |
| Not done | 7 | 5 | 4 | 5 |
| Total | 47 | 34 (72.3%) | 22 (46.8%) | 34 |
Abbreviation: not done, G-/Q-banding analysis was not performed.
Of 26 patients with normal karyotype, metaphase and interphase DCFISH detected translocation of the IgH gene in 9 (34.6%) and 17 (65.4%), respectively (Table 5). Of 7 patients whose cytogenetic information was unavailable because of lack of metaphase spreads, 5 showed split signals of the IgH gene. Interphase FISH also detected the split in 10 (47.6%) of 21 patients who did not have translocation of the IgH gene as assessed by metaphase FISH analysis (Table 3).
DISCUSSION
Chromosome 14q32.33 translocation was detected in 34 (72.3%) of 47 patients with MM and related disorders by combined G-banding and DCFISH methods. The incidence observed in MM was extremely high when compared with NHL and chronic lymphocytic leukemia studied by the same approach.44,51 Previous studies have described that the incidence of 14q32.33 translocatin ranged from 10% to 80% of cytogenetically abnormal cases, with a median of 35%.1-16,52 Although some reports have emphasized that structural abnormalities of chromosome 1 are more common than 14q+ chromosome,2,3,7,10,11,12,16,52 the present study confirms our cytogenetic studies showing the involvement of chromosome 14q32.3 in 62% of patients with abnormal karyotypes.18 Our fingings indicate that translocation of the IgH gene is implicated in the most early molecular and pathogenetic event during the developmental process of MM.
Using DCFISH with YAC and bacteriophage clones, we assessed signals of the IgH gene on metaphase spreads in 43 (91.5%) patients and on interphase nuclei in 46 (97.9%) patients. More analyzable cells were obtained from FISH than G-banding study because obvious signals were shown even on fuzzy or condensed chromosomes. DCFISH with VH and Cγ probes was particularly useful for demonstrating translocation of the IgH gene in patients with chromosomal abnormalities other than 14q32.33 translocations (9 of 11 patients [81.8%]) or in those with normal karyotypes (17 of 26 patients [65.4%]). In both cases, interphase FISH was superior to metaphase FISH in sensitivity to detect this characteristic translocation. Thus, DCFISH with the IgH gene probes is a highly reproducible procedure for detecting 14q32.33 translocations in MM. In some cases, the frequency of the cells with 14q32.33 translocation was lower than expected from the high percentage of myeloma cells in the BM and PB. This discrepancy is explained by the criteria that confined cells with YGR, YYG, YYR, GR, and YR patterns to be positive for the IgH gene translocation and/or by the contamination of PB to heparinized BM samples.
In 11 (50%) of 22 patients with split signals on metaphase spreads, we identified the 14q32.33 translocations specific to the subtype of NHL. Chromosome t(11; 14)(q13.3; q32.33) was found most frequently (5 of 22 patients [22.7%]), followed by t(8; 14)(q24; q32) (2 of 22 [9.1%]) and t(14; 18) (q32.33; q21.3) (2 of 22 [9.1%]; Table 4). A complex 14q32.33 translocation involving 3q and 16q24 was detected in patient no. 2. Signals of VH and Cγ gene probes were observed on der(3)t(3; 14)(q25; q23) and der(16)t(15; 16) (q11;q24), respectively. This alteration seems to occur stepwisely as a result of complex chromosomal translocations during the development of MM. Alternatively, Cγ may be inserted into the breakpoint at band 16q24 of der(16) t(15; 16).
We identified 7q32.1 and 16q24 as novel partners of 14q32.33 tranlocations in MM. Previous studies showed that donor sites of 14q32.33 translocations in MM were 4p16.3, 6p21.1, 8q24, 11q13, and 18q21 recurrently, and 1q21, 3p11, 7q11.2, 11q23, and 16q23.1 singly.18,21 Bergsagel et al21 have recently cloned various breakpoints of 14q32.33 translocation, including 4p16.3, 8q24.13, 16q23.1, and 21q22.1 in MM cell lines. Among these subsets of 14q32.33 translocation, it was reported that t(11; 14) frequently occurs in myeloma with morphologic features characteristic of small cleaved plasma cells16 and that t(8; 14) is associated with an IgA isotype.9 In our study, t(7; 14)(q32.1; q32.3) was found in a patient with myeloma cells expressing myelomonocytoid antigen. A previous report has documented a structural abnormality involving band 7q32.1 in a MM cell line with myelomonocytoid feature.53 Our findings suggest that a number of patients with MM not only share a similar pathogenetic background with B-cell NHL, but also have a specific genetic event for MM.
Chromosome t(11; 14)(q13.3; q32.33) was detected in 5 (10.6%) of 47 patients as assessed by DCFISH. Only 1 of these patients had MM, whereas 3 had PCL and 1 had plasmacytoma. In the previous study, t(11; 14) was detected in 10% of patients successfully analyzed by conventional banding method.18 This translocation was observed in 3% of patients in the series demonstrated by Dewald et al6 and 4% of patients studied by 6-day culture method with granulocyte-macrophage colony-stimulating factor, interleukin-3, and/or interleukin-6 in the series reported by Lai et al.13
The translocation, t(8; 14)(q24.1; q32.33), was detected in 2 patients by combined DCFISH and G-banding. A variant t(8; 22)(q24.1; q11) was also detected in 2 patients by G-banding. These were confirmed by DCFISH with painting probes for chromosome 8, 14, and 22 (data not shown). The findings suggest that c-MYC is involved in 4 (9.5%) of 42 patients with MM and plasma cell malignancies. One of the 2 patients with t(8; 14) showed split signals of whitish pink (corresponding to Y) and pink (corresponding to R) through triple-band-pass filter, each of which was observed on 14q+ and donor chromosome at 8q24.1, respectively. The results suggested rearrangement within the VH region covered by Y6. VH gene segments were rarely involved in 14q32.33 translocations of B-cell malignancies, which have only been documented in a cell line derived from Burkitt's lymphoma with t(8; 14).54
Chromosome t(14; 18)(q32.33; q21.3) was found in 2 patients, as confirmed by DCFISH with BCL2 and VH gene probes. We have previously reported 2 patients with MM carrying this translocation18,36 and detected 2 additional patients in the present study. The association between t(14; 18) and MM has been established by the chromosomal analysis and bcl-2 protein expression analysis.9 55
DCFISH identified 14q32.33 translocations in 3 (60%) of 5 patients with MGUS and in 14 (60.9%) of 23 patients with plasma cell malignancies carrying normal karyotype. Our findings showed genetic rearrangements of plasma cells in more than half of patients with MGUS and MM with normal karyotypes in bone marrow cells, suggesting malignant proliferation of BM plasma cells (BMPC) in these disorders. Although a significant increase of the monoclonal protein or development to myeloma, macroglobulinemia, or amyloidosis occurred in approximately 20% of patients with MGUS, little was known about the pathophysiologic mechanisms.56 Cytogenetically abnormal clones were rarely found in BMPC of patients with MGUS. More recently, the incidence of aneuploidy of chromosomes 3, 7, 11, and 18 was lower in MGUS than MM as assessed by FISH.57 In contrast, the incidence of split within the IgH gene in MGUS appears to be similar to MM. Based on the clinical and cytogenetic observations, we propose that 14q32.33 translocation is a primary change in the pathogenesis of MGUS. Hence, DCFISH with using the IgH gene probes is a promising approach to elucidate the molecular mechanisms leading to abnormal proliferation and differentiation in MGUS, smoldering myeloma, and asymptomatic myeloma, as well as MM.
In conclusion, we demonstrated high incidence of translocations of the IgH gene in MM as detected by DCFISH with Cγ and VH gene probes. The method was rapid and sensitive in screening the IgH gene translocation on metaphase spreads and interphase nuclei of MM and related disorders at a single-cell level. According to the donor sites of 14q32.33 translocations identified, it would be possible not only to classify MM but also to isolate novel proto-oncogenes specific to MM.
ACKNOWLEDGMENT
The authors are grateful to Prof Tasuku Honjo (Center for Molecular Biology and Genetics, Kyoto University, Kyoto, Japan), Dr Teruyuki Takashima, Dr Shigeo Horiike, and Dr Shinichi Misawa for valuable discussions. We thank Dr Gary A. Silverman (Harvard Medical School, Boston, MA) for providing the YAC clone yA153A6. We thank the following doctors for their kind supply of samples and their clinical data: Drs Motoharu Kondo and Hiroyuki Kobayashi (First Department of Internal Medicine, Kyoto Prefectual University of Medicine, Kyoto, Japan); Drs Masao Nakagawa and Chihiro Shimazaki (Second Department of Internal Medicine, Kyoto Prefecutural University of Medicine); Drs Haruo Sugiyama and Yasuo Ogawa (Third Department of Internal Medicine, Osaka University, Osaka, Japan); Drs Shouichiro Tsuda and Yasuo Ogawara (Aiseikai-Yamashina Hospital, Kyoto, Japan); Dr Hiroyuki Nakai (Otsu City Hospital, Otsu, Japan); Mitsuru Tsudo (Katsura Hospital, Kyoto, Japan); Drs Hiroshi Fujii and Hitoshi Nakagawa (Kyoto First Red Cross Hospital, Kyoto, Japan); Dr Shouhei Yokota (Yosanoumi Hospital, Kyoto, Japan); and Kenjiro Hamamoto (Kishiwada City Hospital, Osaka, Japan).
Supported in part by Grants-in-Aid from the Ministry of Education (Grant No. 05151048) and the Ministry of Health and Welfare, Japan.
Address reprint requests to Masafumi Taniwaki, MD, Third Department of Internal Medicine, Kyoto Prefectural University of Medicine, Kawaramachi-Hirokoji, Kamigyo-ku, Kyoto 602, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal